
M
E
T
A
Z
O
O
G
E
N
E

|
P L A N K T O N B A R C O D E A T L A S & D A T A B A S E
|
Data Access Page
Within MZGdb, each taxonomic group and geographic region has its own Data Access page, allowing a user to download only the taxonomic group or region of interest.
MZGdb offers the data in FASTA format, along with a paired taxonomic hierarchy file (semicolon-separated, Mothur compatible), as well as "the MZGdb format", in comma-separated-values (CSV) and pipe-separated-values (PSV) format.
(The CSV file should load into Excel, and other CSV-capable programs.
The PSV file, which uses a pipe "|" character instead of a comma to separate columns, is a legacy/back-up format mainly used for debugging.
There is a challenge in making a working/legal CSV file out of the GenBank and BOLD records, which themselves use commas (and other weird characters) in their own data values.)
The full MZGdb csv/psv format is described here (click for info on the MZGdb Format).
The "Mode-A" / "Mode-B" / "Mode-C" / "Mode-D" download options (previously discussed in the Group Summary) determine whether barcodes sampled from outside of the current region are included in the data set (and if Genus-only data are included), which in turn determines how many barcodes (and their species coverage) will be downloaded.
-
The "Mode-A" data set focuses on included as many species as possible, even when the barcodes come from other oceans or regions.
This option is suggested for people looking for an all-inclusive reference data set that can identify as many species as possible.
- - - > In most cases, you will want to download and use the "mode-A" MZGdb data set.
For maximum barcode matches, you also probably want to use the "world" (o00) files.
-
The "Mode-B" data set only included species barcodes sampled from the current ocean or region.
This option is suggested for people that want to only look at "local" barcodes to reduce geographic variations.
Mode-B may greatly reduce the amount of barcodes (and the number species barcoded) available, however.
-
"Mode-C / Mode-D" results are not shown in this table. "Mode-C" data is any-ocean data like "Mode-A", but it can include sequence records only identified to Genus level.
"Mode-D" is specific-ocean data like "Mode-B", but likewise include Genus-only sequence records.
The "Mode-C / Mode-D" data can also contain uncertain species identifications like "Calanus aff. finmarchicus".
In general, the best species and barcode coverage is available via the "Mode-A" data files.
The other "Mode" files are designed for more custom or special-focus applications.
Two Cautions about "Mode-A" and "Mode-B" data options:
-
Using multiple different regions of "Mode-A" / "Mode-C" data (Duplication):
Because Mode-A can include species barcodes from other oceans/regions, combining MZGdb data files from two or more regions (e.g. combining the North Atlantic and the Arctic) will often cause duplicated barcode records.
Depending on your analysis software, this may cause issues, or they may just be flagged and ignored.
If you want to do multiple regions, and this causes problem with your software, use the entire-world data set, as that will contain all regions but have no duplicates.
(The "Mode-B" data does not, by definition and design, have duplicates between regions.)
-
"Mode-B" / "Mode-D" Data Sparsity:
The map below shows observation locations of Neocalanus gracilis (blue dots) and the locations of geo-referenced barcodes for this species (red starts).
In "Mode-B", barcodes for this species would only be present in the Mode-B North Pacific and Mode-B South Pacific data sets.
Likewise, "Mode-B" data from the Indian Ocean or North Atlantic would not include this species.
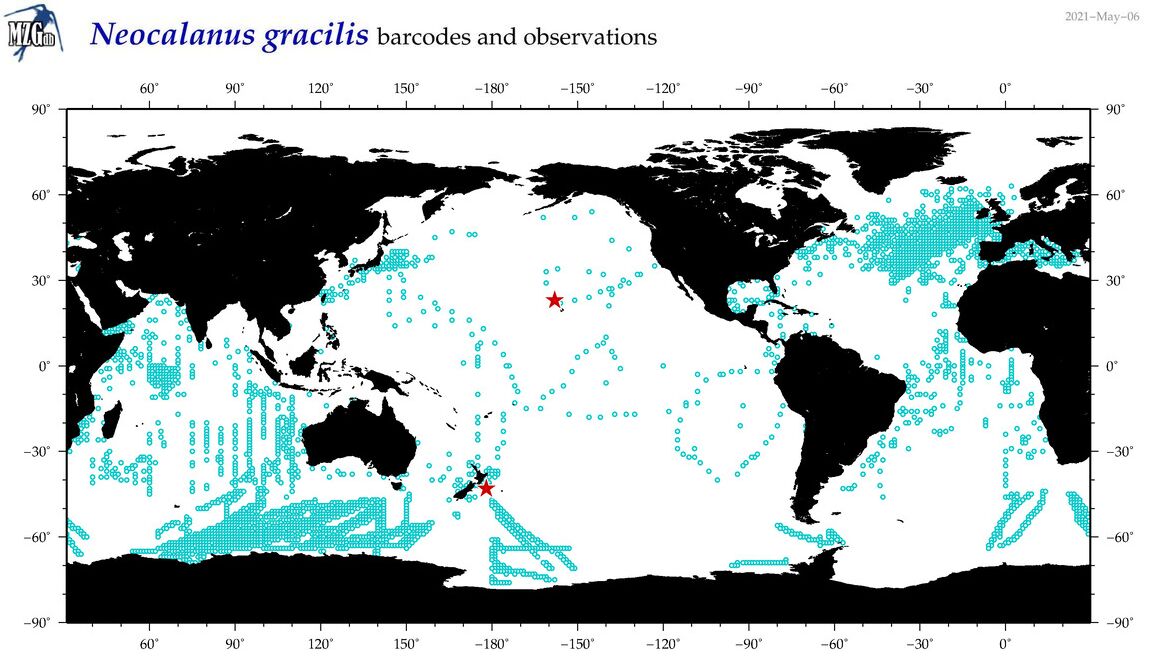
Example of a globally-present (copepod) species with far-from-global barcoding coverage.
|
|
|