Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2023-m07-15
2023-Jul-20 |
| Abyssobrotula galatheae |
Species
(1) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5015835
R:1:0:0:0 |
| Acanthonus armatus |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
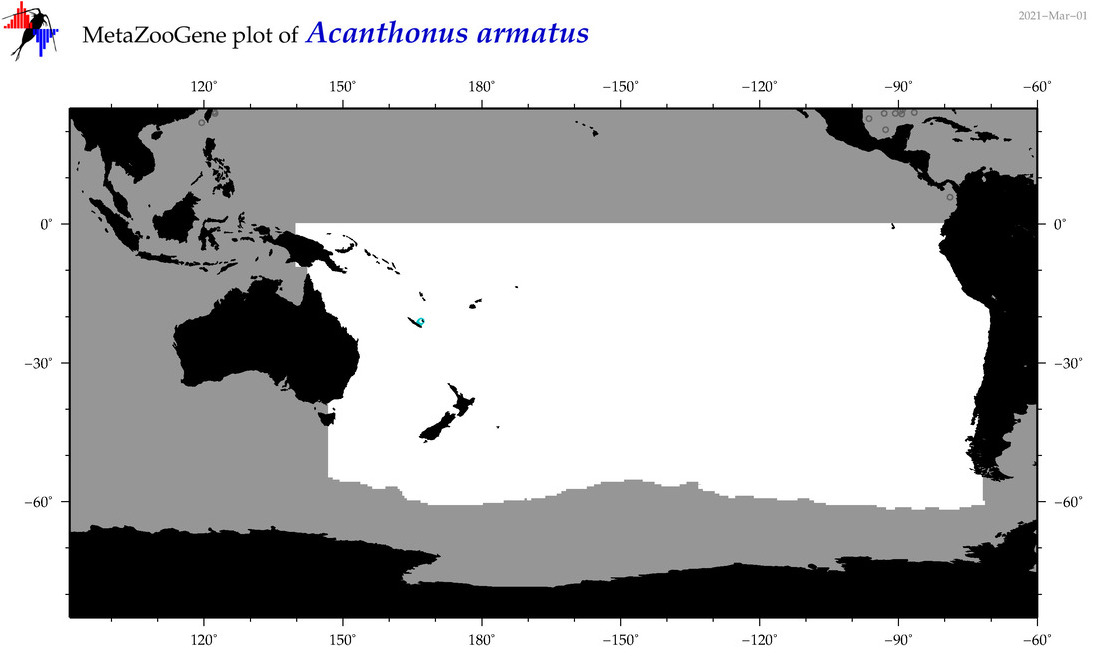 |
accepted
T5003029
R:1:0:0:0 |
| Alionematichthys crassiceps |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027673
R:1:0:0:0 |
| Alionematichthys piger |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
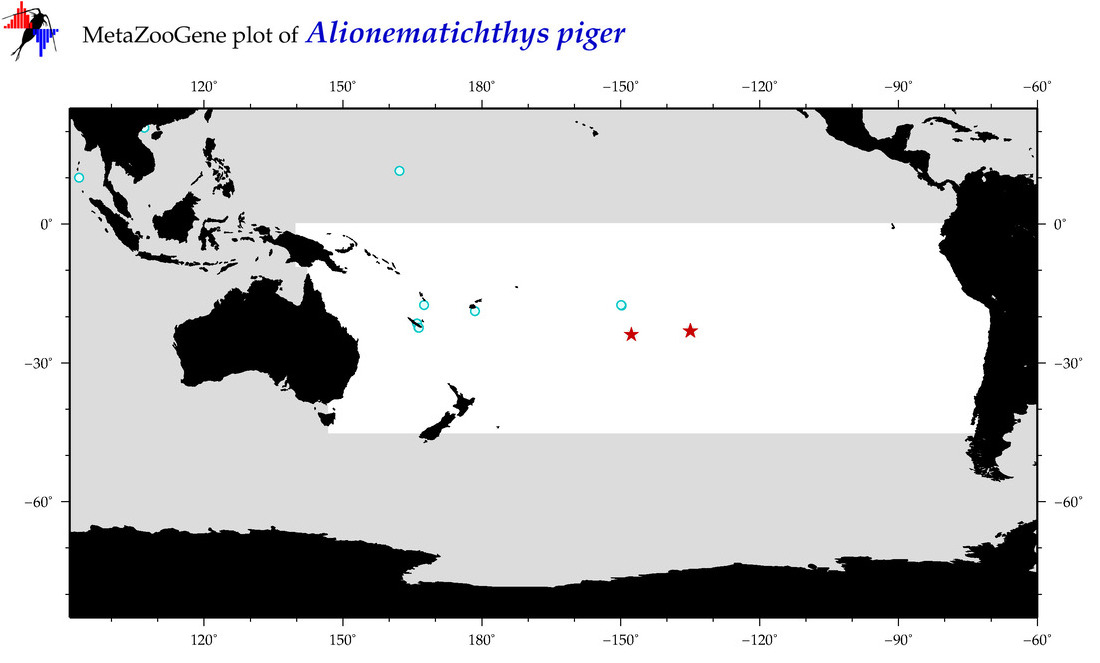 |
accepted
T5003060
R:1:0:0:0 |
| Alionematichthys plicatosurculus |
Species
(5) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016053
R:1:0:0:0 |
| Alionematichthys riukiuensis |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016054
R:1:0:0:0 |
| Alionematichthys winterbottomi |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016056
R:1:0:0:0 |
| Aphyonus bolini |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016376
R:1:0:0:0 |
| Aphyonus gelatinosus |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
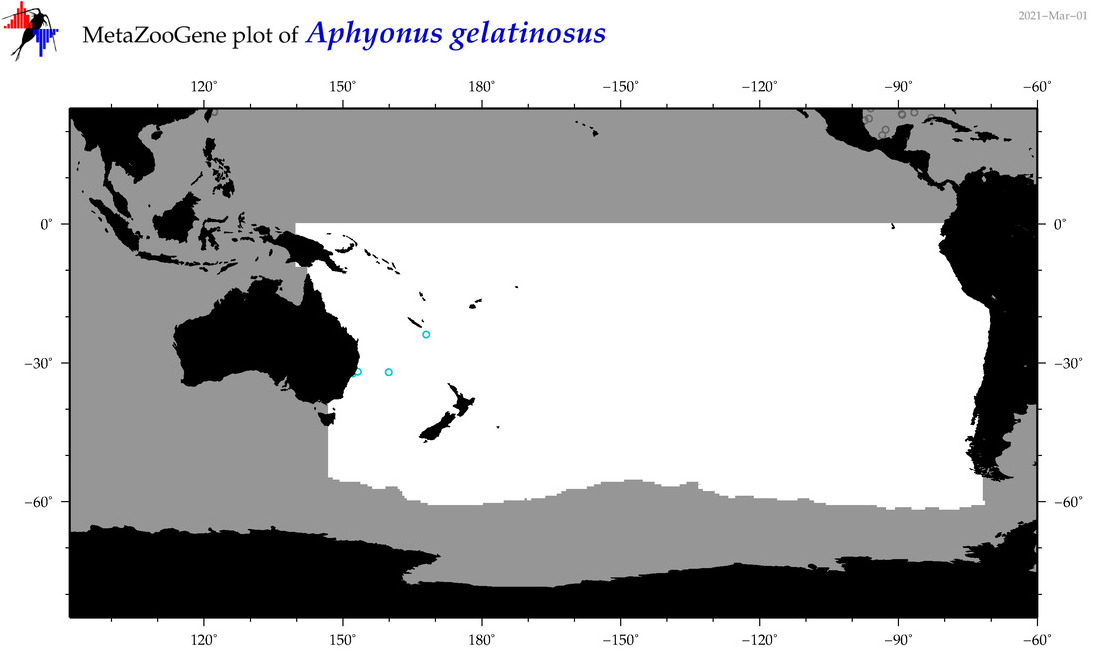 |
accepted
T5007842
R:1:0:0:0 |
| Barathronus maculatus |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
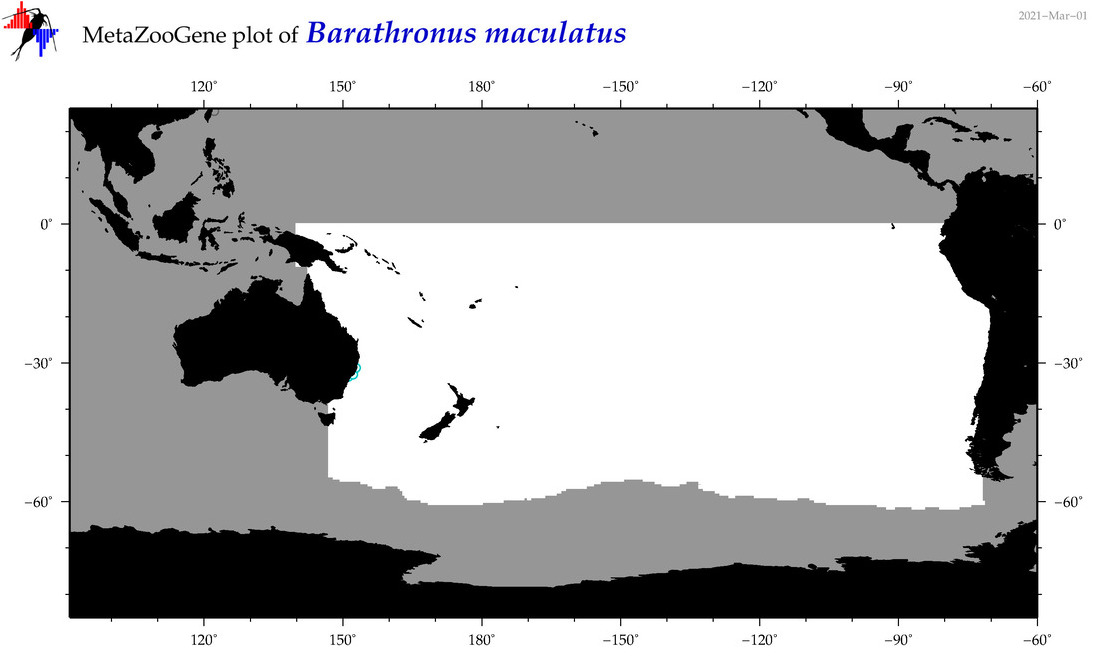 |
accepted
T5008064
R:1:0:0:0 |
| Bassogigas gillii |
Species
(11) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016963
R:1:0:0:0 |
| Bassozetus elongatus |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016964
R:1:0:0:0 |
| Bassozetus glutinosus |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
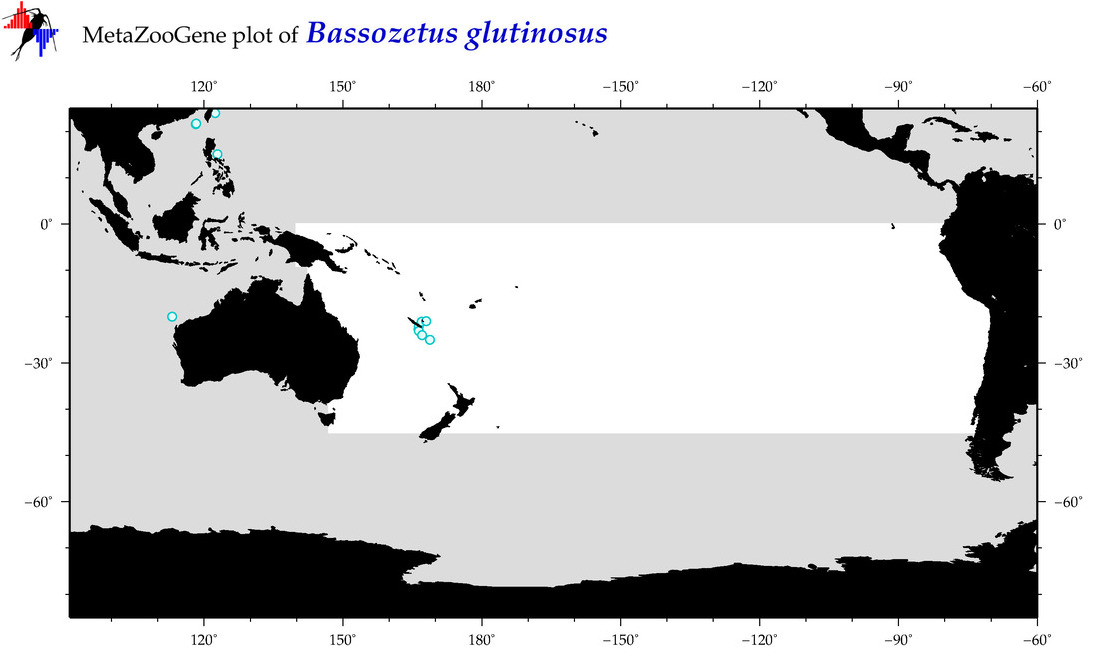 |
accepted
T5003237
R:1:0:0:0 |
| Bassozetus robustus |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016970
R:1:0:0:0 |
| Bathyonus caudalis |
Species
(15) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017052
R:1:0:0:0 |
| Bidenichthys beeblebroxi |
Species
(16) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017165
R:1:0:0:0 |
| Bidenichthys consobrinus |
Species
(17) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027769
R:1:0:0:0 |
| Brosmophyciops pautzkei |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
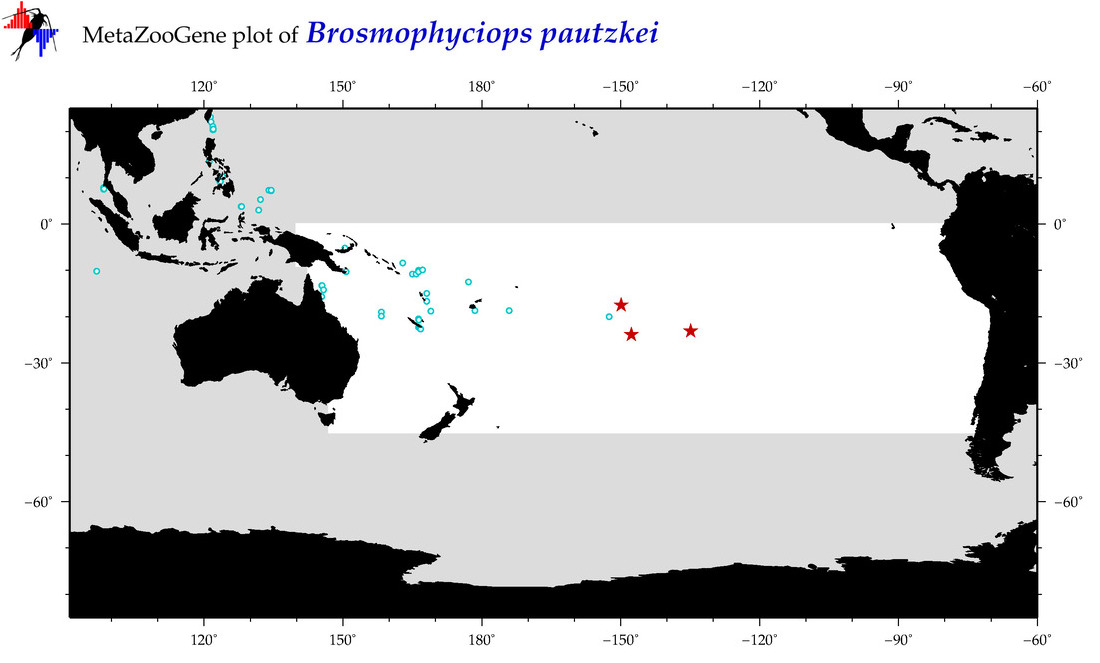 |
accepted
T5003315
R:1:0:0:0 |
| Brotula clarkae |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 51
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
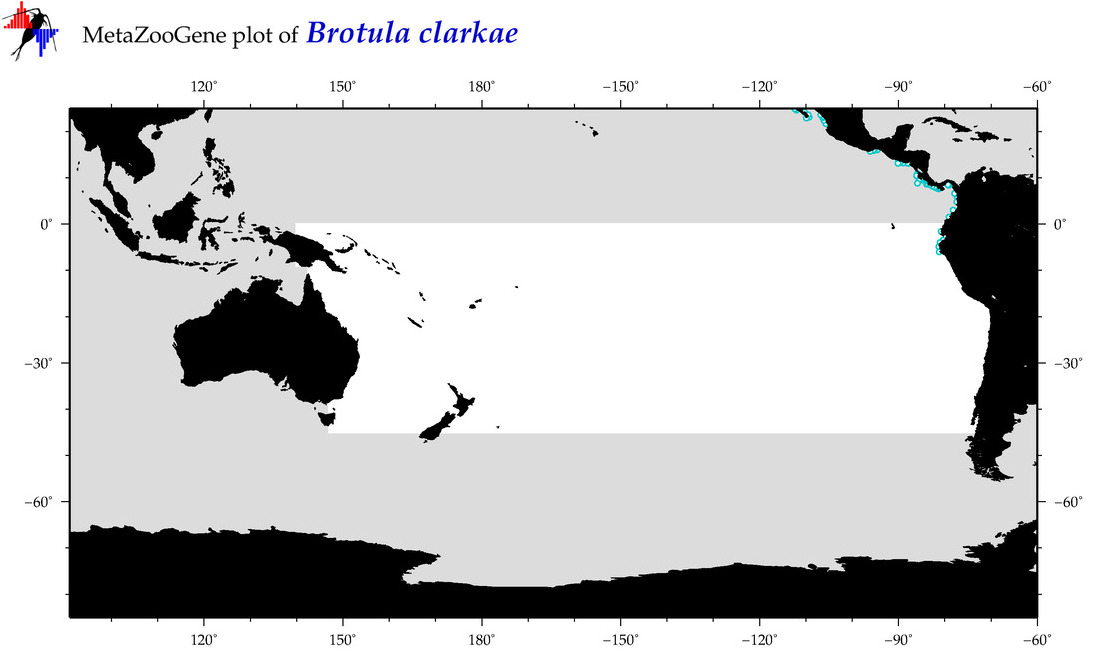 |
accepted
T5001839
R:1:0:0:0 |
| Brotula multibarbata |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
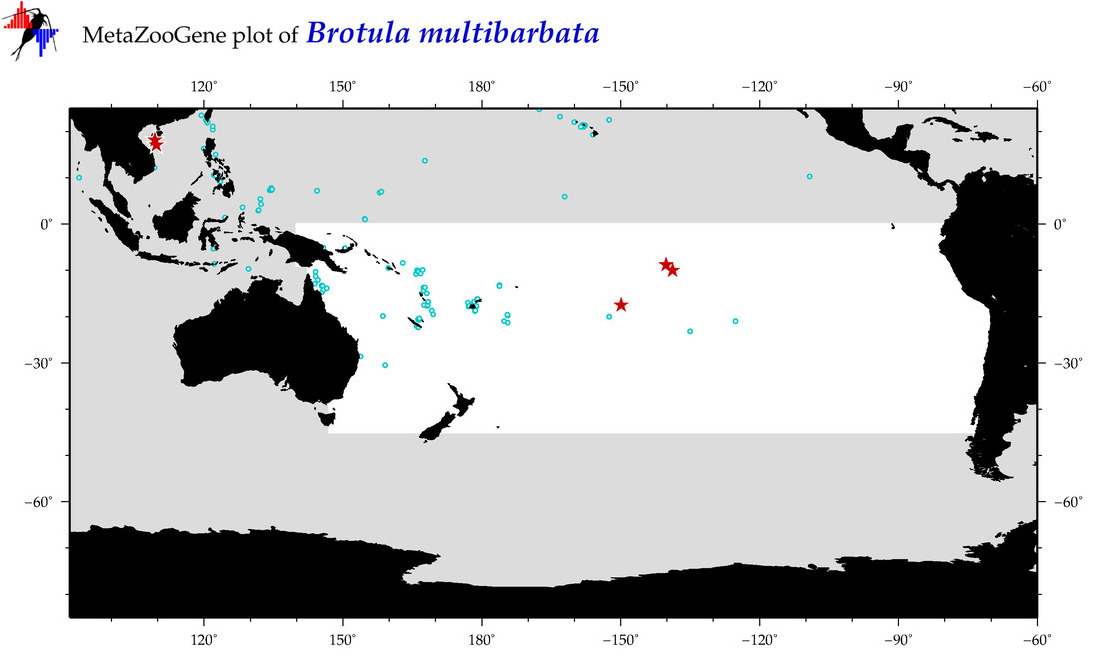 |
accepted
T5003317
R:1:0:0:0 |
| Brotula ordwayi |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
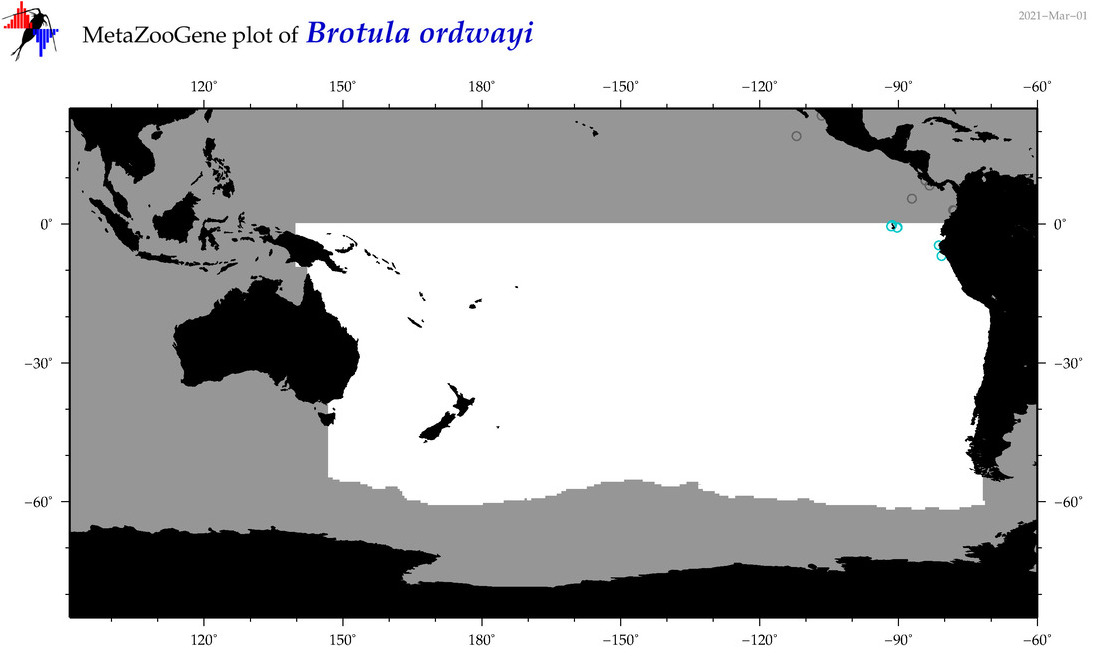 |
accepted
T5017344
R:1:0:0:0 |
| Brotula townsendi |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
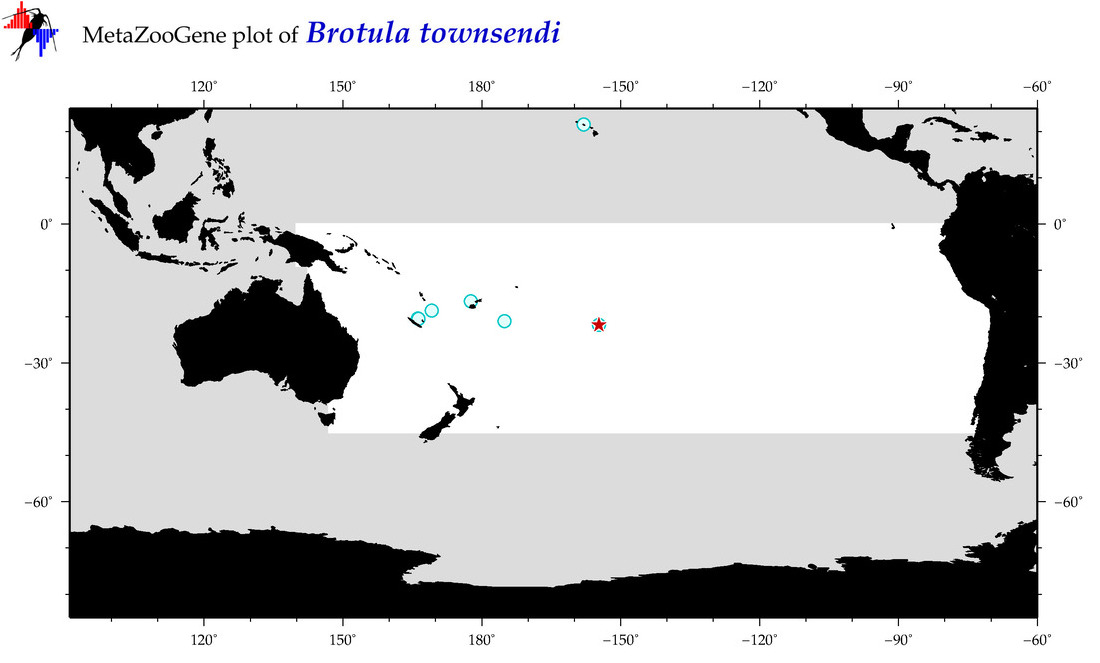 |
accepted
T5003318
R:1:0:0:0 |
| Brotulotaenia crassa |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
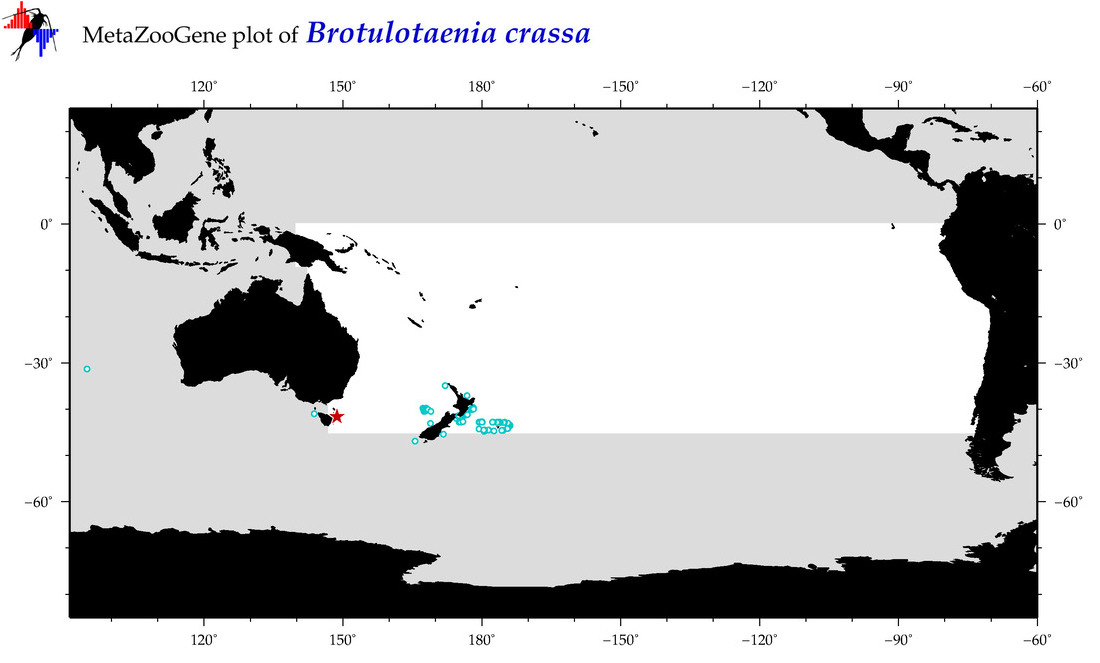 |
accepted
T5008218
R:1:0:0:0 |
| Brotulotaenia nigra |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
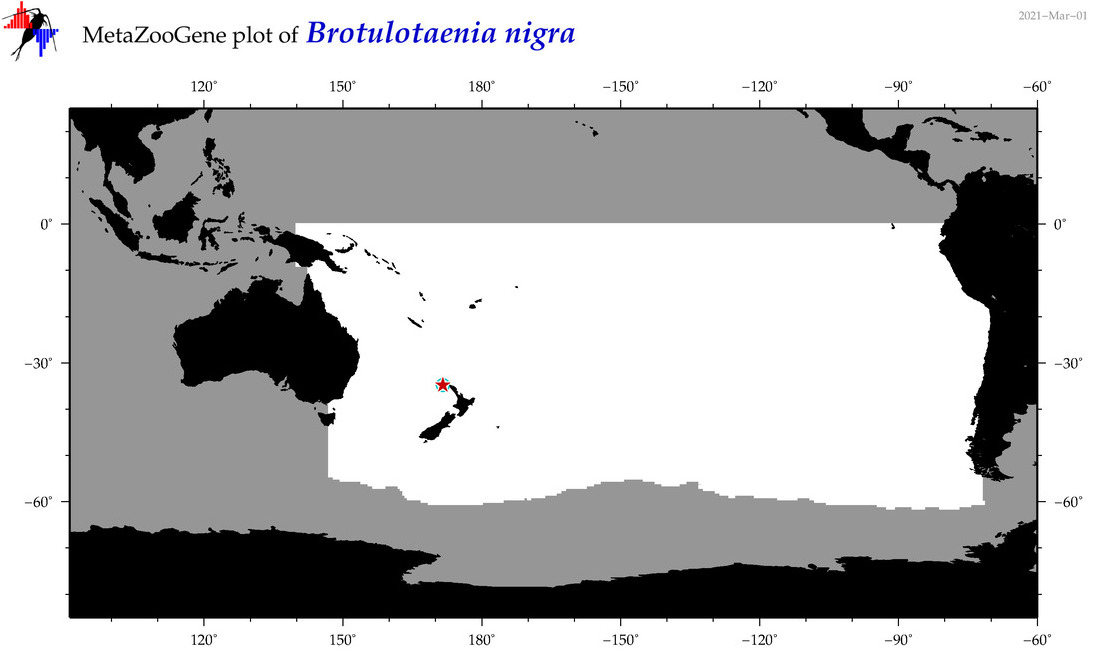 |
accepted
T5008219
R:1:0:0:0 |
| Calamopteryx jeb |
Species
(25) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017421
R:1:0:0:0 |
| Carapus mourlani |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 5
16S = 3
18S = 2
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
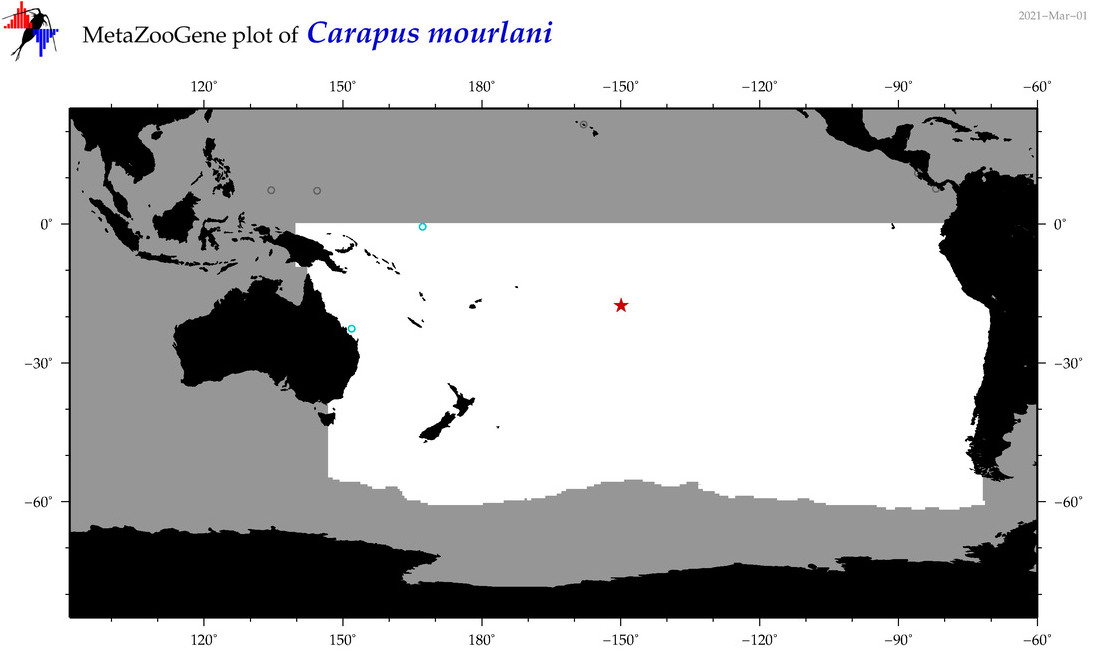 |
accepted
T5003385
R:1:0:0:0 |
| Cataetyx chthamalorhynchus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
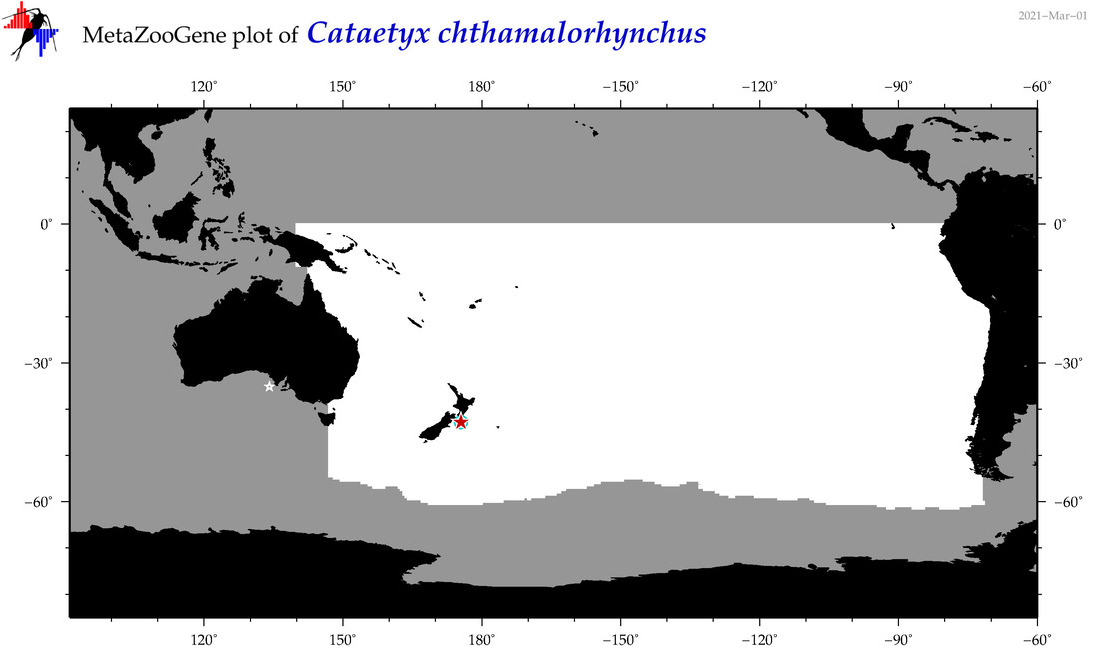 |
accepted
T5003418
R:1:0:0:0 |
| Cataetyx nielseni |
Species
(28) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027830
R:1:0:0:0 |
| Cataetyx niki |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
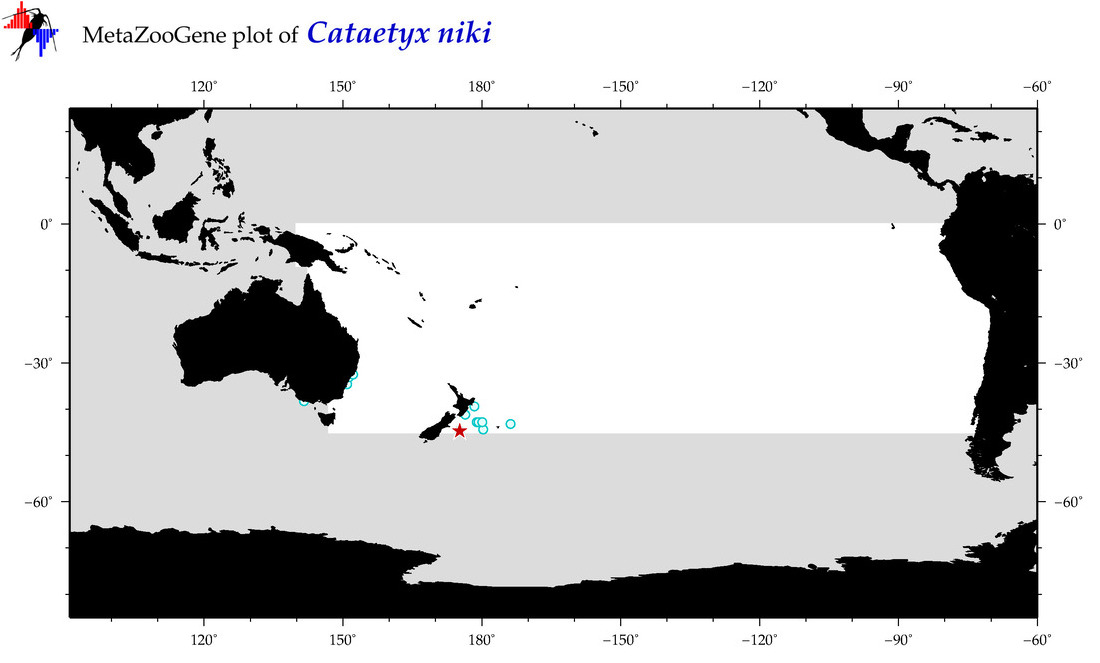 |
accepted
T5003420
R:1:0:0:0 |
| Cataetyx rubrirostris |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 2
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
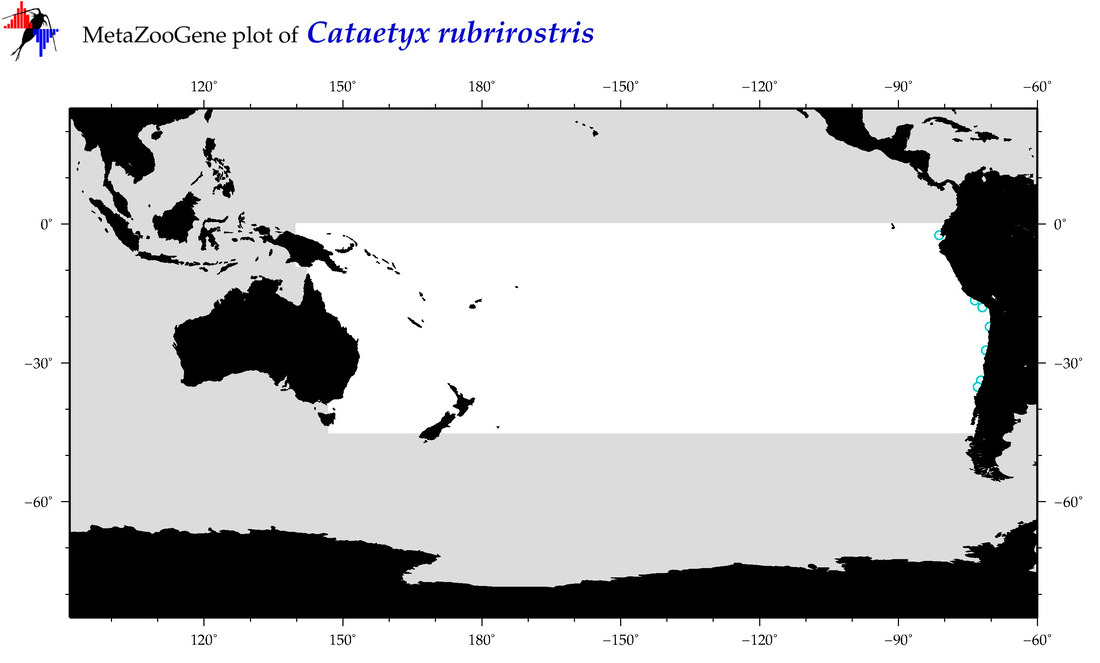 |
accepted
T5000349
R:1:0:0:0 |
| Cataetyx simus |
Species
(31) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017654
R:1:0:0:0 |
| Cherublemma emmelas |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
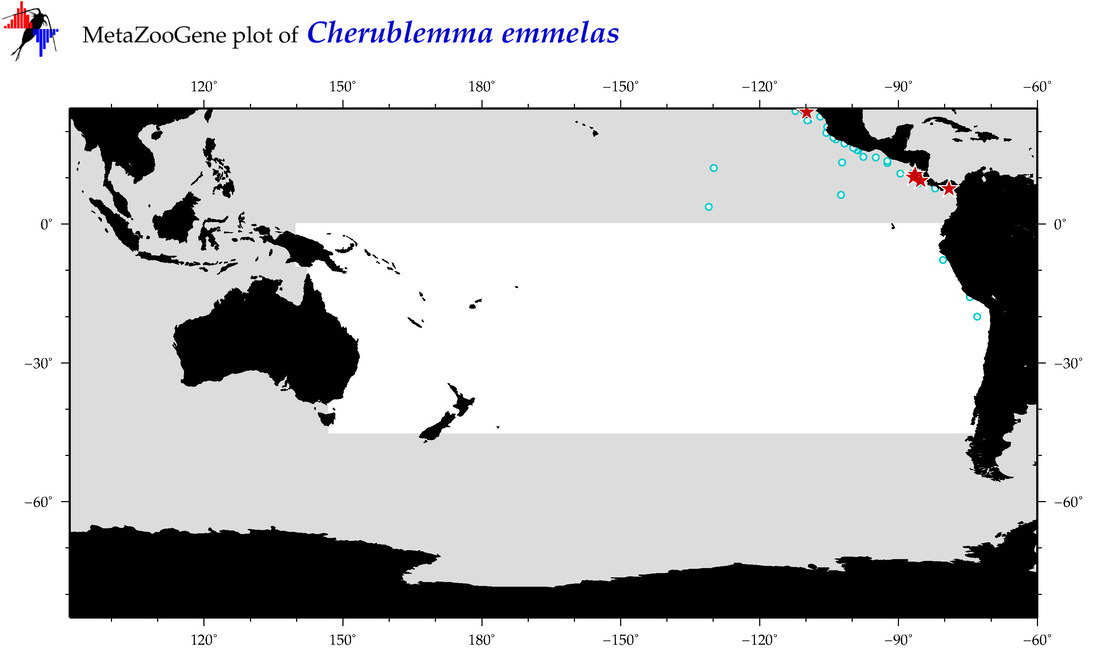 |
accepted
T5000388
R:1:0:0:0 |
| Dannevigia tusca |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018965
R:1:0:0:0 |
| Dermatopsis greenfieldi |
Species
(34) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019038
R:1:0:0:0 |
| Dermatopsis hoesei |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019039
R:1:0:0:0 |
| Dermatopsis macrodon |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019041
R:1:0:0:0 |
| Diancistrus alleni |
Species
(37) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019057
R:1:0:0:0 |
| Diancistrus altidorsalis |
Species
(38) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019058
R:1:0:0:0 |
| Diancistrus beateae |
Species
(39) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019059
R:1:0:0:0 |
| Diancistrus brevirostris |
Species
(40) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019060
R:1:0:0:0 |
| Diancistrus eremitus |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019061
R:1:0:0:0 |
| Diancistrus fijiensis |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019063
R:1:0:0:0 |
| Diancistrus katrineae |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
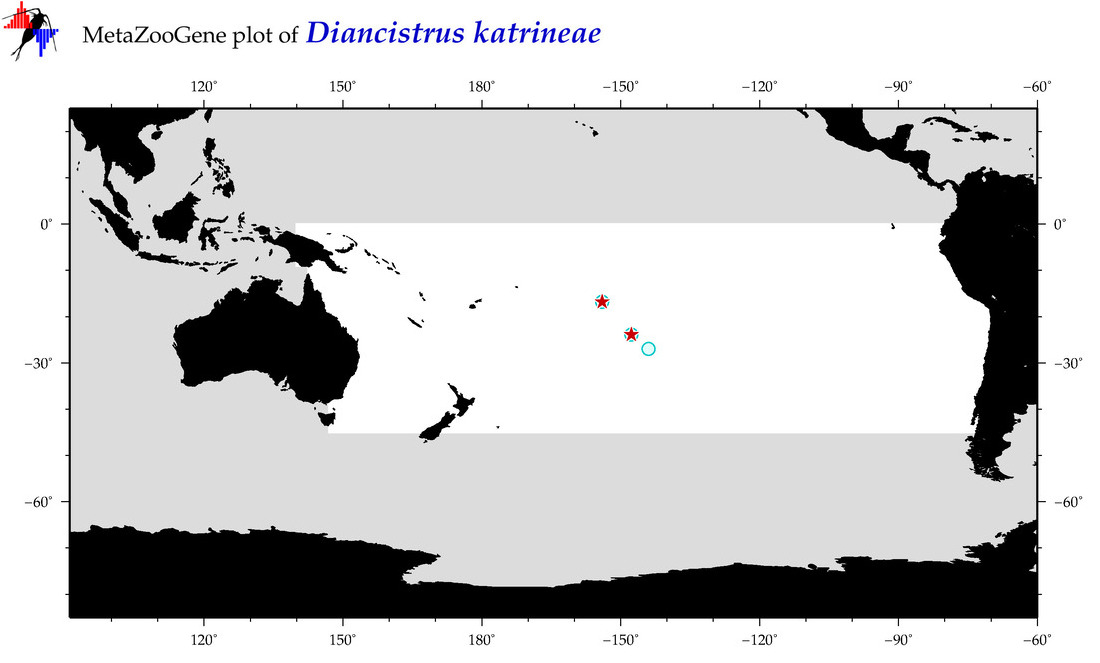 |
accepted
T5003749
R:1:0:0:0 |
| Diancistrus leisi |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019066
R:1:0:0:0 |
| Diancistrus longifilis |
Species
(45) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019067
R:1:0:0:0 |
| Diancistrus mcgroutheri |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019070
R:1:0:0:0 |
| Diancistrus mennei |
Species
(47) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027966
R:1:0:0:0 |
| Diancistrus novaeguineae |
Species
(48) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019072
R:1:0:0:0 |
| Diancistrus tongaensis |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019076
R:1:0:0:0 |
| Dicrolene filamentosa |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
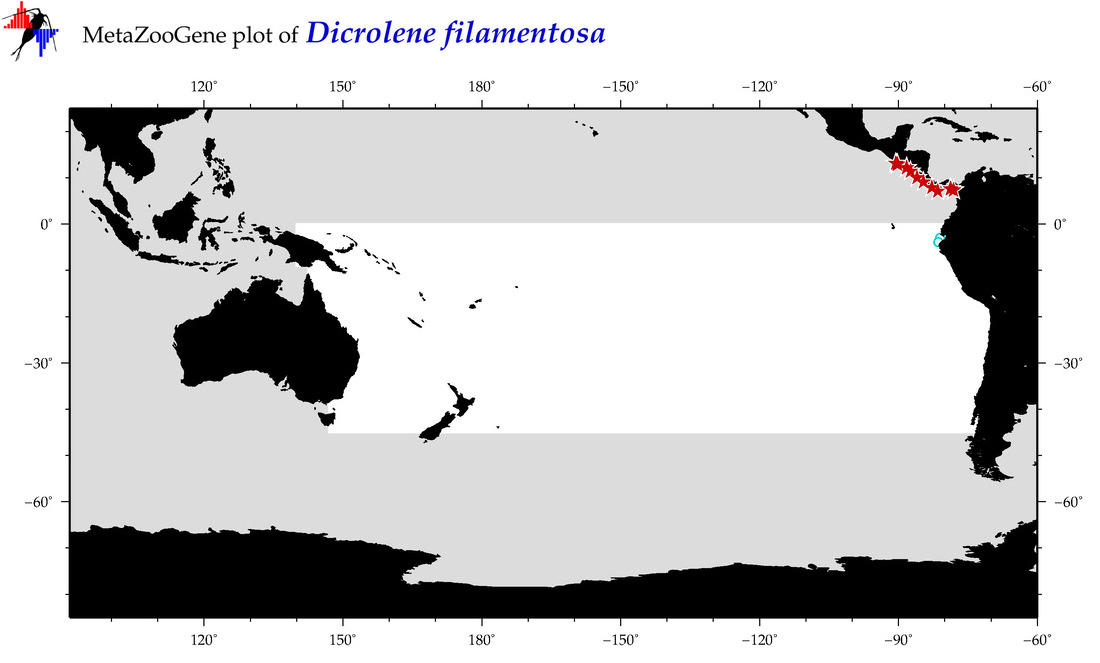 |
accepted
T5003785
R:1:0:0:0 |
| Dicrolene introniger |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
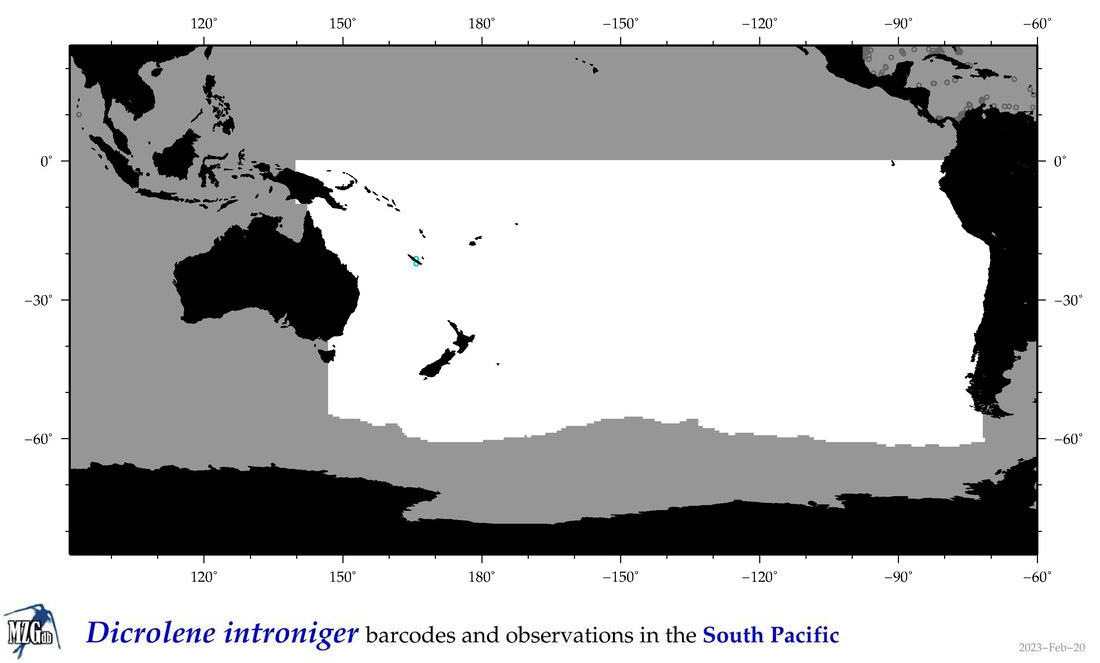 |
accepted
T5003786
R:1:0:0:0 |
| Dicrolene longimana |
Species
(52) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019133
R:1:0:0:0 |
| Dicrolene nigra |
Species
(53) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019135
R:1:0:0:0 |
| Didymothallus criniceps |
Species
(54) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027975
R:1:0:0:0 |
| Didymothallus mizolepis |
Species
(55) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027976
R:1:0:0:0 |
| Didymothallus pruvosti |
Species
(56) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027978
R:1:0:0:0 |
| Dinematichthys iluocoeteoides |
Species
(57) |
COI
12S
16S
18S
28S
|
COI = 19
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
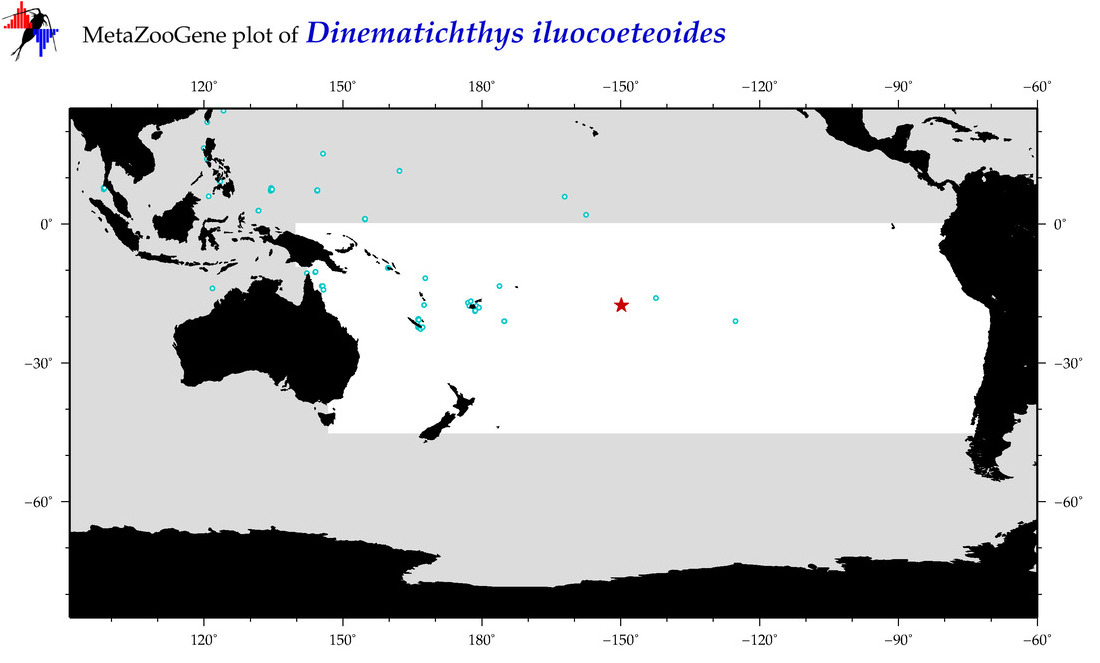 |
accepted
T5003791
R:1:0:0:0 |
| Diplacanthopoma brachysoma |
Species
(58) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019146
R:1:0:0:0 |
| Diplacanthopoma jordani |
Species
(59) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019148
R:1:0:0:0 |
| Dipulus norfolkanus |
Species
(60) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019195
R:1:0:0:0 |
| Echiodon anchipterus |
Species
(61) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019263
R:1:0:0:0 |
| Echiodon cryomargarites |
Species
(62) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
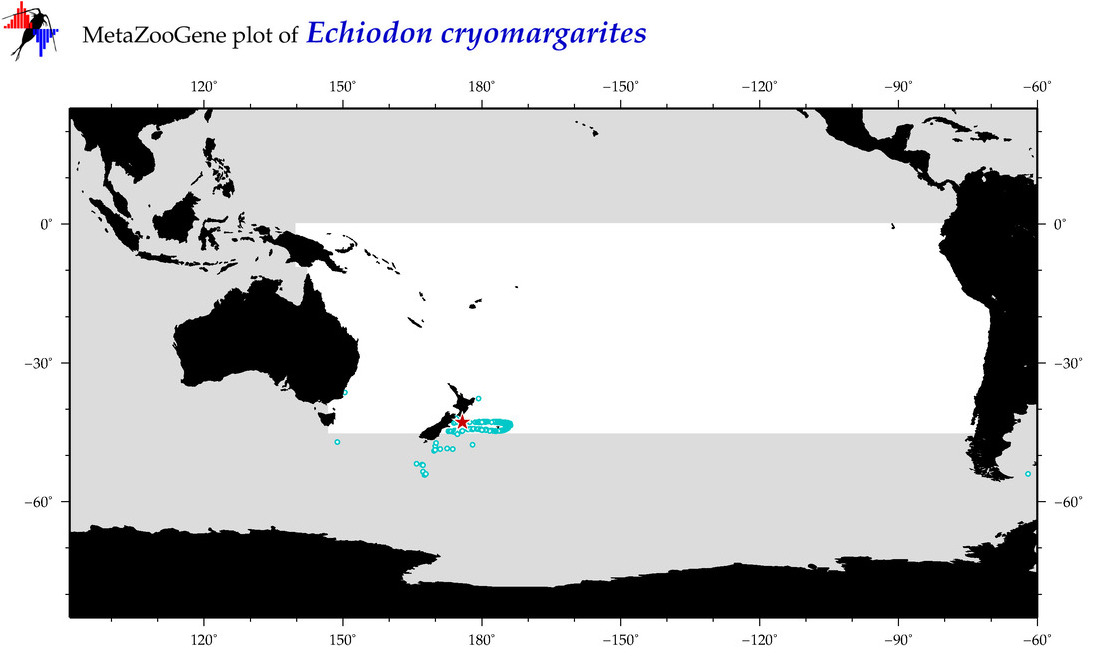 |
accepted
T5003842
R:1:0:0:0 |
| Echiodon exsilium |
Species
(63) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5000526
R:1:0:0:0 |
| Echiodon rendahli |
Species
(64) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019267
R:1:0:0:0 |
| Ematops randalli |
Species
(65) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019388
R:1:0:0:0 |
| Encheliophis boraborensis |
Species
(66) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027998
R:1:0:0:0 |
| Encheliophis chardewalli |
Species
(67) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027999
R:1:0:0:0 |
| Encheliophis gracilis |
Species
(68) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 5
16S = 4
18S = 3
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019420
R:1:0:0:0 |
| Encheliophis homei |
Species
(69) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 4
16S = 4
18S = 3
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019421
R:1:0:0:0 |
| Encheliophis vermicularis |
Species
(70) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019422
R:1:0:0:0 |
| Eurypleuron cinereum |
Species
(71) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019703
R:1:0:0:0 |
| Eurypleuron owasianum |
Species
(72) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019704
R:1:0:0:0 |
| Eusurculus pistillum |
Species
(73) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028031
R:1:0:0:0 |
| Genypterus blacodes |
Species
(74) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 3
16S = 7
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
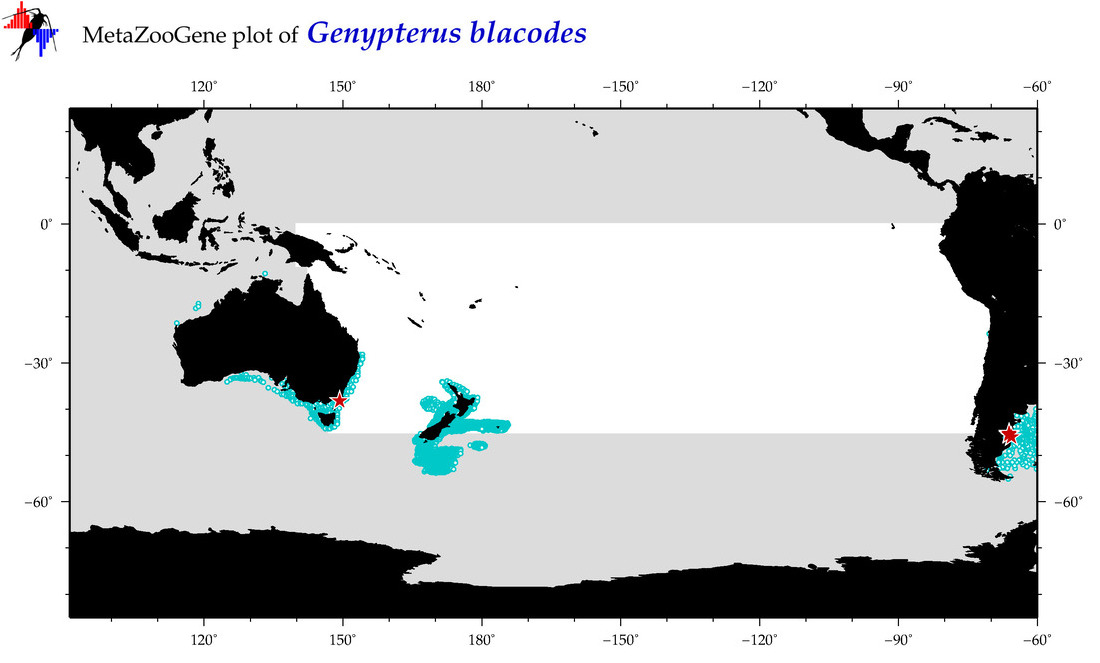 |
accepted
T5004053
R:1:0:0:0 |
| Genypterus maculatus |
Species
(75) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020062
R:1:0:0:0 |
| Genypterus tigerinus |
Species
(76) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
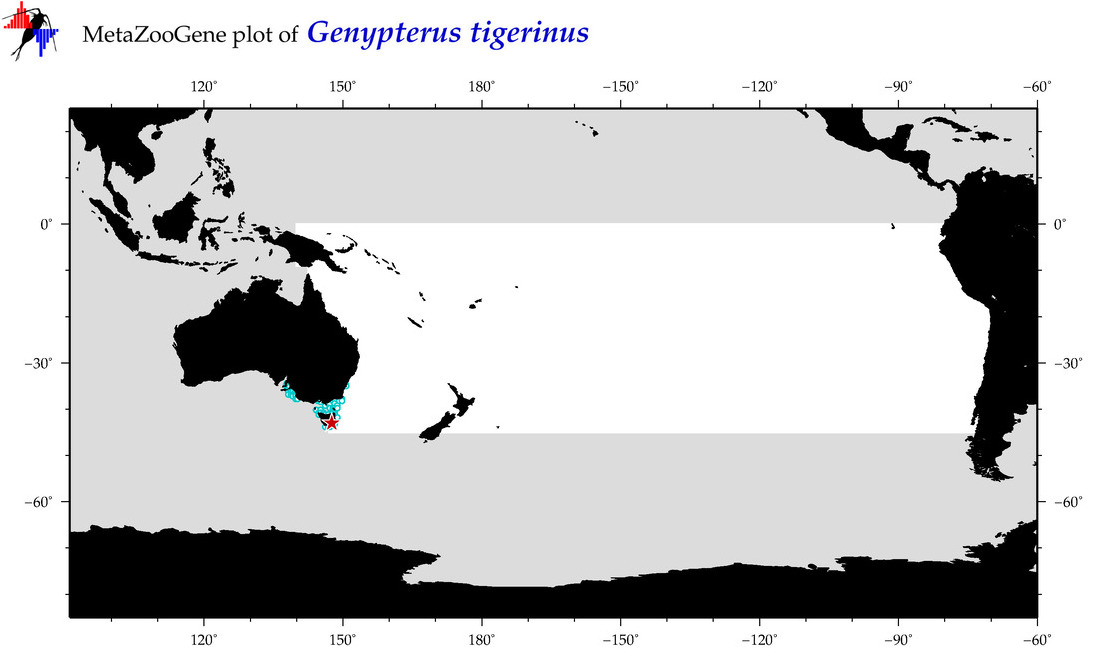 |
accepted
T5004056
R:1:1:0:0 |
| Glyptophidium argenteum |
Species
(77) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020164
R:1:0:0:0 |
| Glyptophidium japonicum |
Species
(78) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020166
R:1:0:0:0 |
| Grammonus diagrammus |
Species
(79) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020337
R:1:0:0:0 |
| Grammonus robustus |
Species
(80) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020339
R:1:0:0:0 |
| Homostolus acer |
Species
(81) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
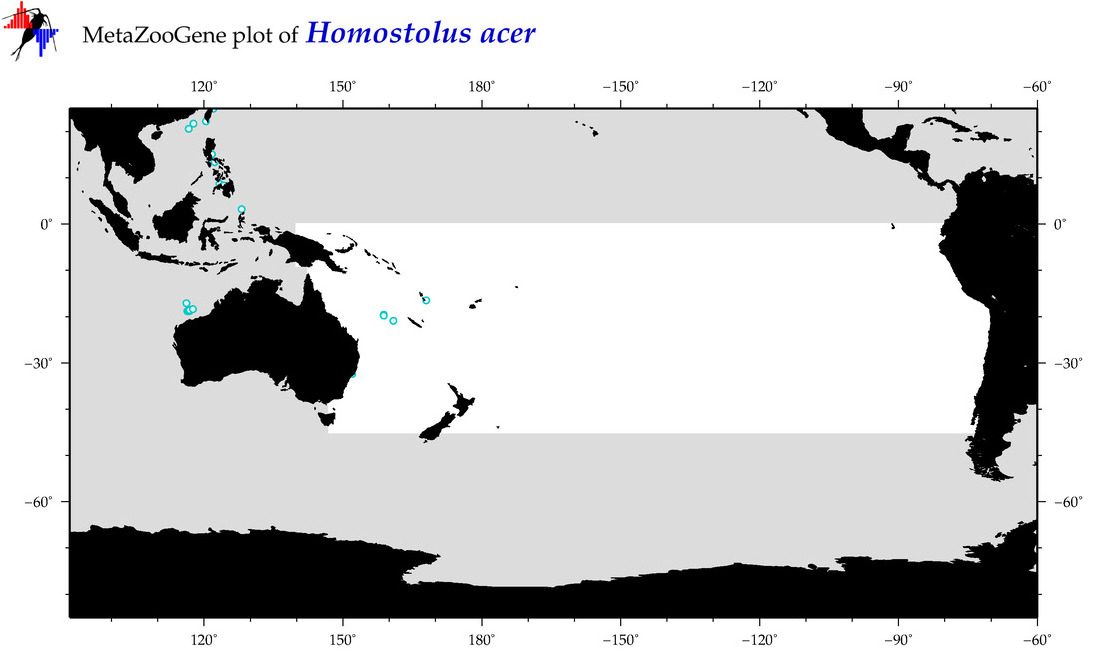 |
accepted
T5004290
R:1:0:0:0 |
| Hoplobrotula armata |
Species
(82) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 6
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
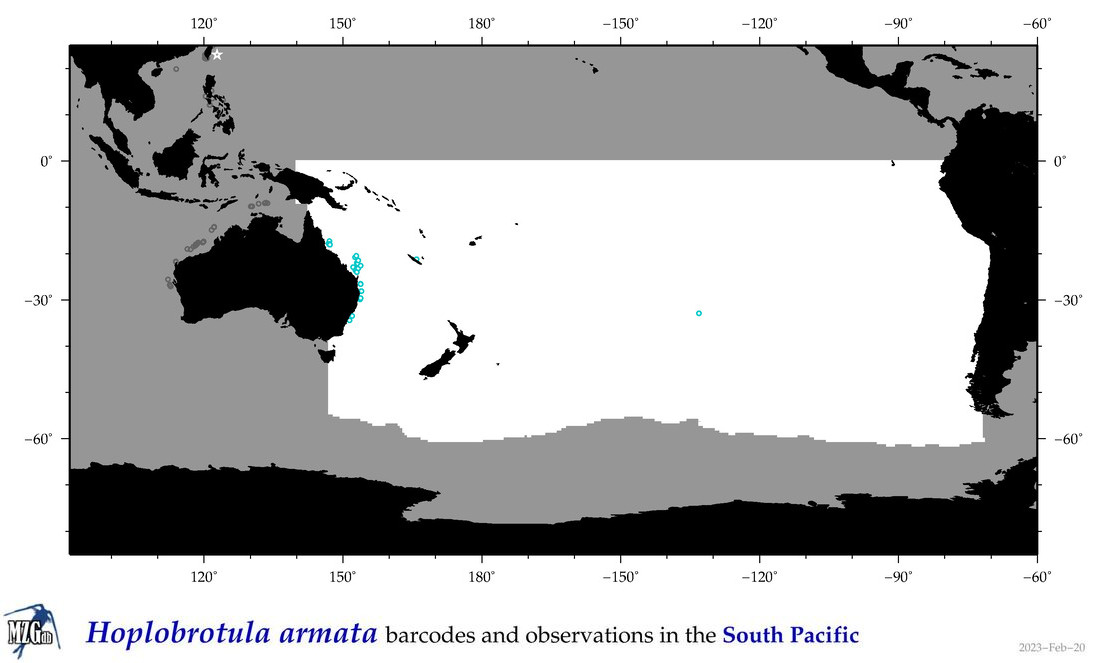 |
accepted
T5004291
R:1:0:0:0 |
| Lamprogrammus niger |
Species
(83) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004420
R:1:0:0:0 |
| Lapitaichthys frickei |
Species
(84) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021393
R:1:0:0:0 |
| Lepophidium brevibarbe |
Species
(85) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
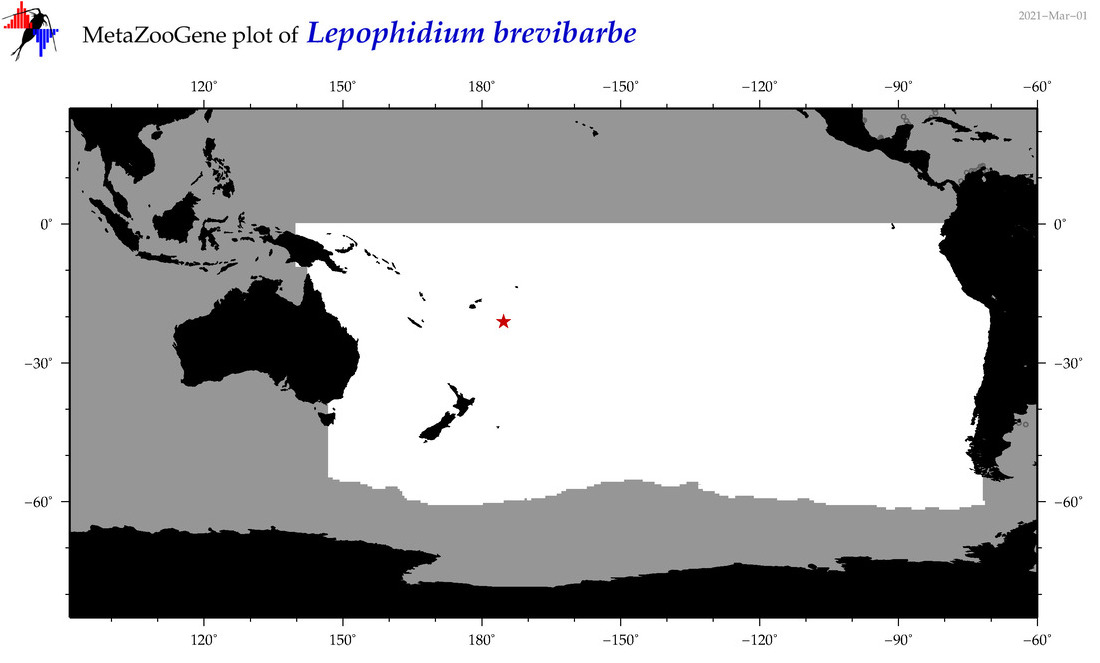 |
accepted
T5004462
R:1:0:0:0 |
| Lepophidium inca |
Species
(86) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028207
R:1:0:0:0 |
| Lepophidium microlepis |
Species
(87) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021516
R:1:0:0:0 |
| Lepophidium negropinna |
Species
(88) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
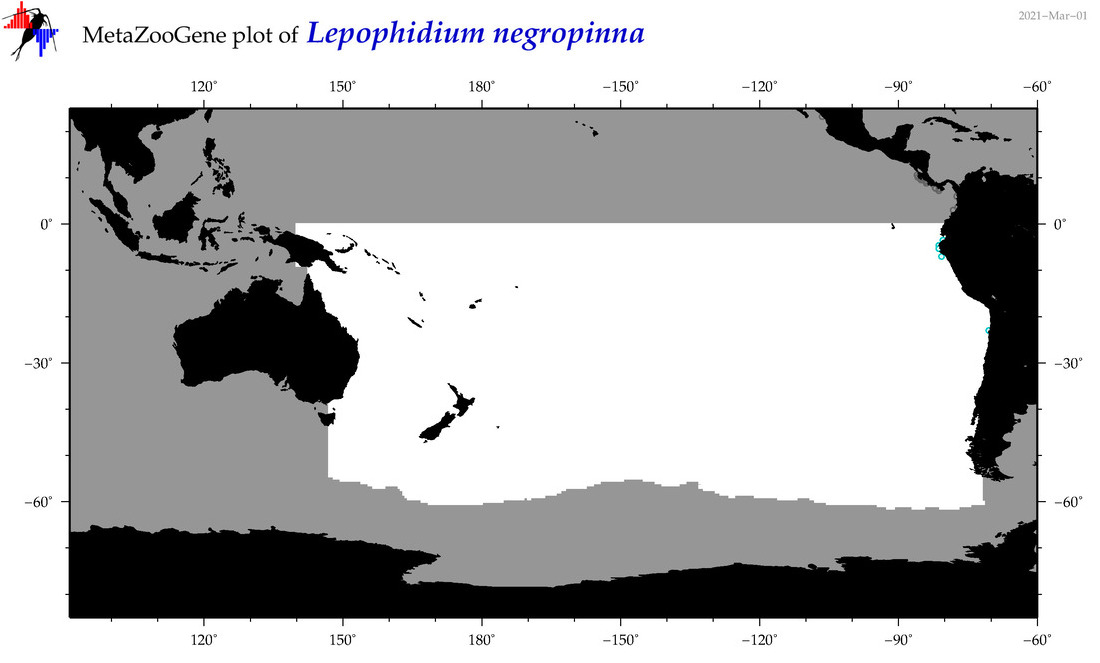 |
accepted
T5000782
R:1:0:0:0 |
| Lepophidium pardale |
Species
(89) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021517
R:1:0:0:0 |
| Lepophidium prorates |
Species
(90) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021519
R:1:0:0:0 |
| Luciobrotula bartschi |
Species
(91) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
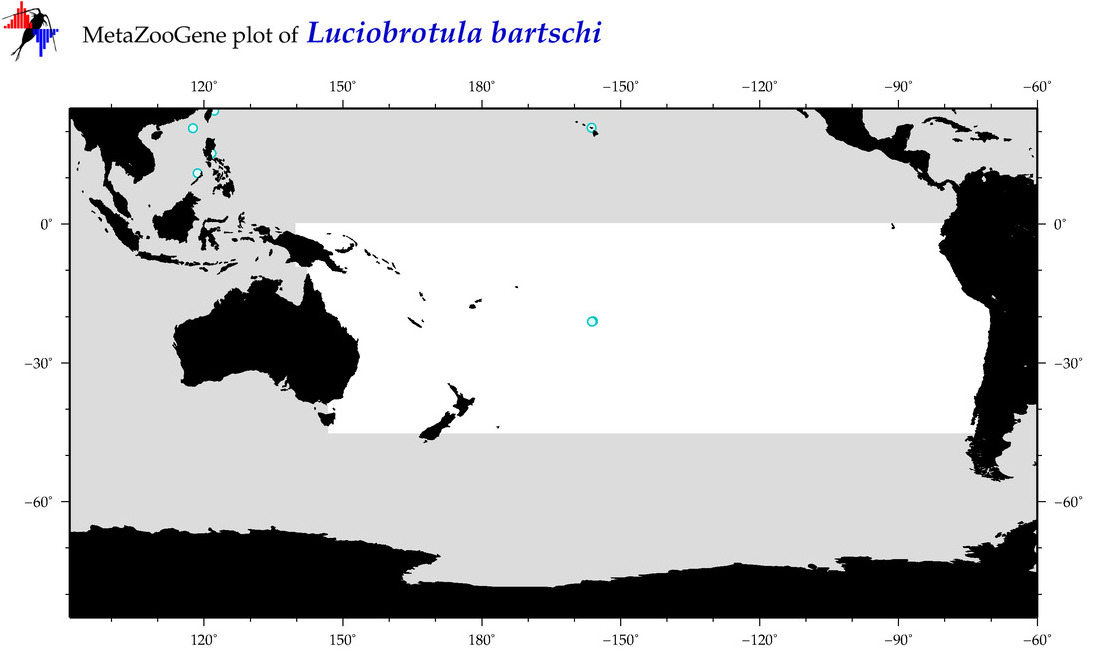 |
accepted
T5010244
R:1:0:0:0 |
| Microbrotula greenfieldi |
Species
(92) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028294
R:1:0:0:0 |
| Microbrotula punicea |
Species
(93) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028296
R:1:0:0:0 |
| Microbrotula queenslandica |
Species
(94) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028297
R:1:0:0:0 |
| Monomitopus agassizii |
Species
(95) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
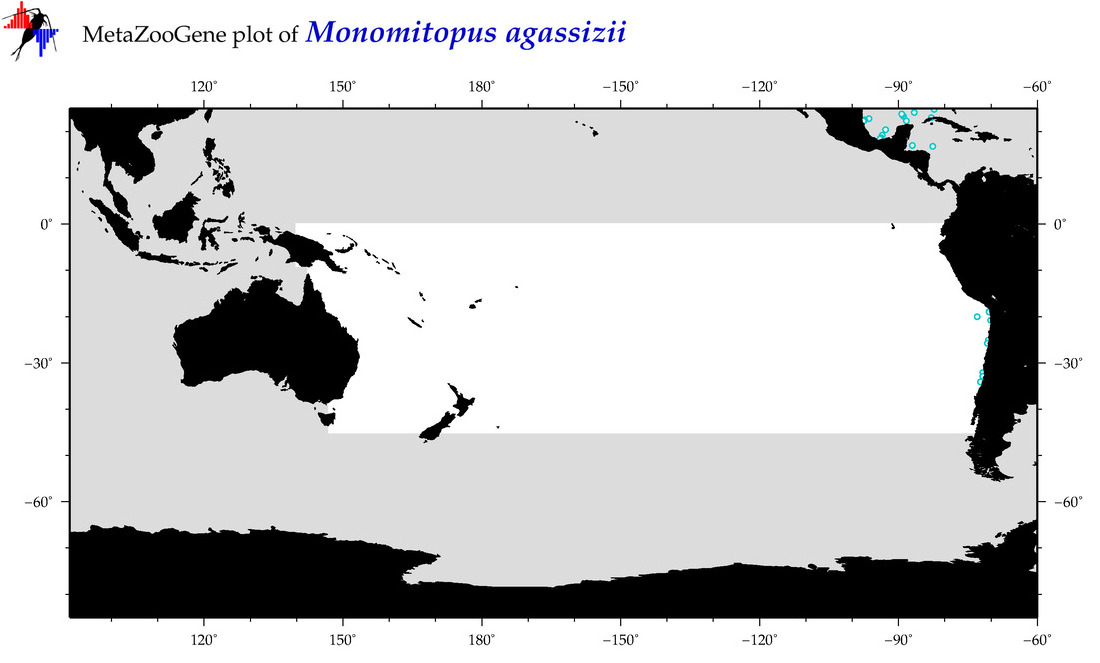 |
accepted
T5004696
R:1:0:0:0 |
| Monomitopus garmani |
Species
(96) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022275
R:1:0:0:0 |
| Monomitopus malispinosus |
Species
(97) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
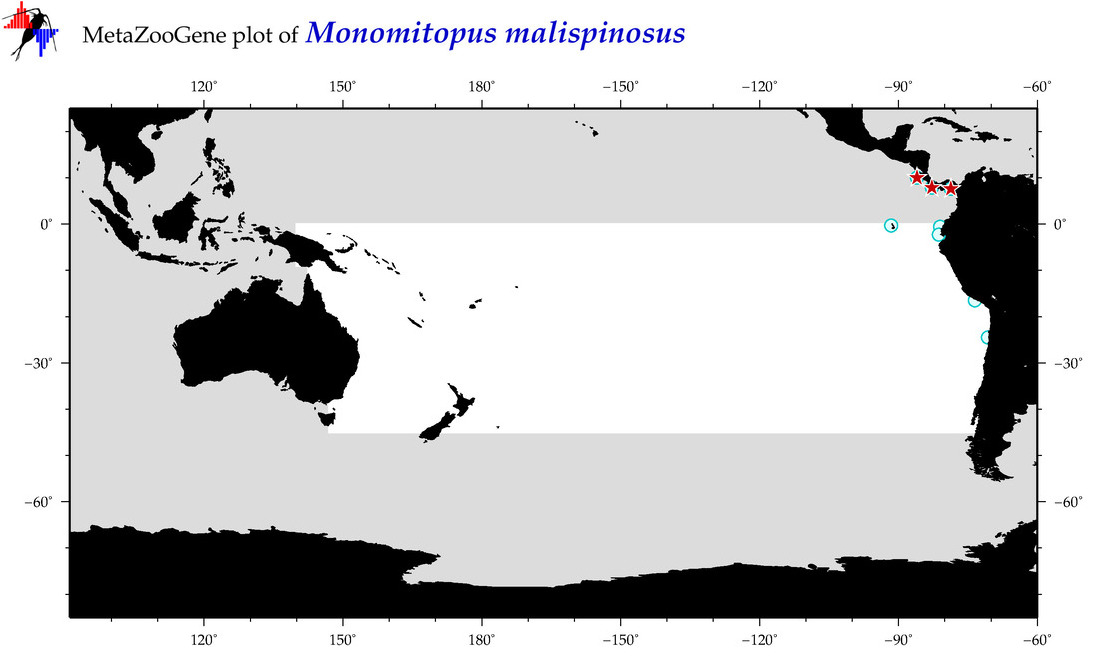 |
accepted
T5004698
R:1:0:0:0 |
| Monomitopus torvus |
Species
(98) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022281
R:1:0:0:0 |
| Monothrix polylepis |
Species
(99) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028312
R:1:0:0:0 |
| Neobythites bimaculatus |
Species
(100) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
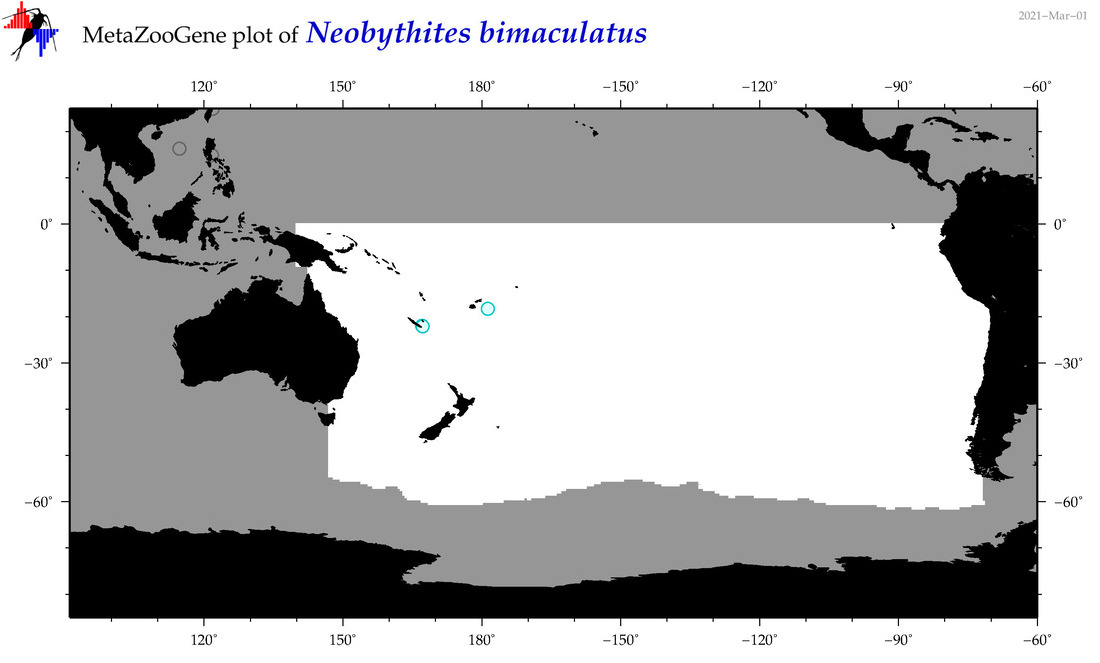 |
accepted
T5010617
R:1:0:0:0 |
| Neobythites bimarginatus |
Species
(101) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
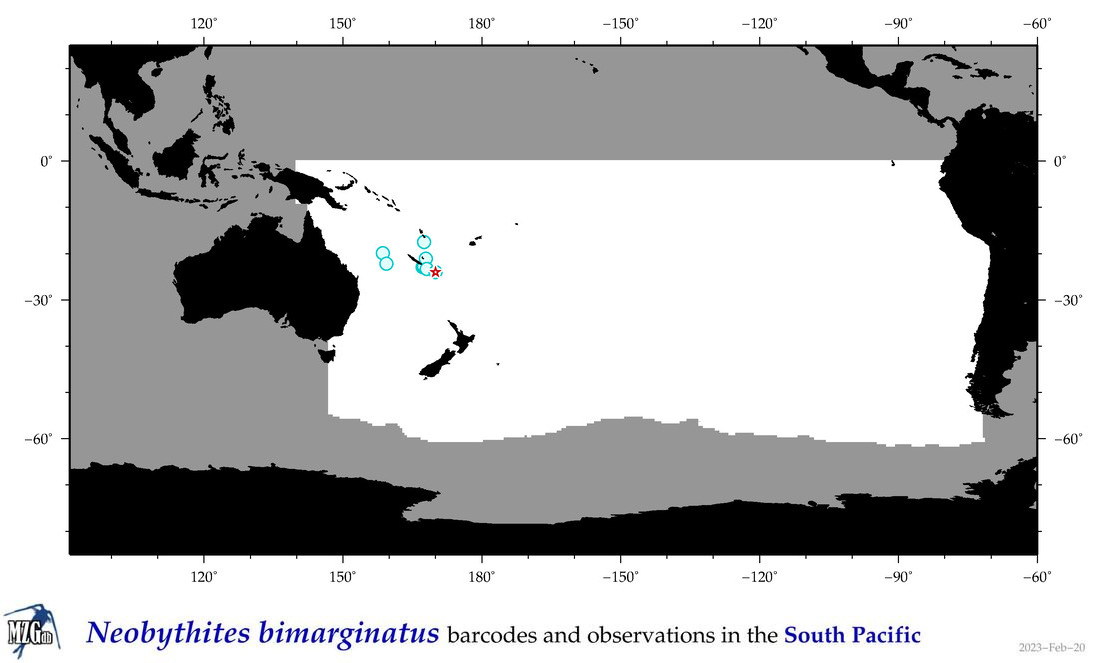 |
accepted
T5022555
R:1:0:0:0 |
| Neobythites franzi |
Species
(102) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022562
R:1:0:0:0 |
| Neobythites longiventralis |
Species
(103) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022567
R:1:0:0:0 |
| Neobythites malayanus |
Species
(104) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022570
R:1:0:0:0 |
| Neobythites marquesaensis |
Species
(105) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022572
R:1:0:0:0 |
| Neobythites neocaledoniensis |
Species
(106) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022577
R:1:0:0:0 |
| Neobythites nigriventris |
Species
(107) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022578
R:1:0:0:0 |
| Neobythites pallidus |
Species
(108) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022580
R:1:0:0:0 |
| Neobythites purus |
Species
(109) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022581
R:1:0:0:0 |
| Neobythites sereti |
Species
(110) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022582
R:1:0:0:0 |
| Neobythites sivicola |
Species
(111) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 5
16S = 3
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
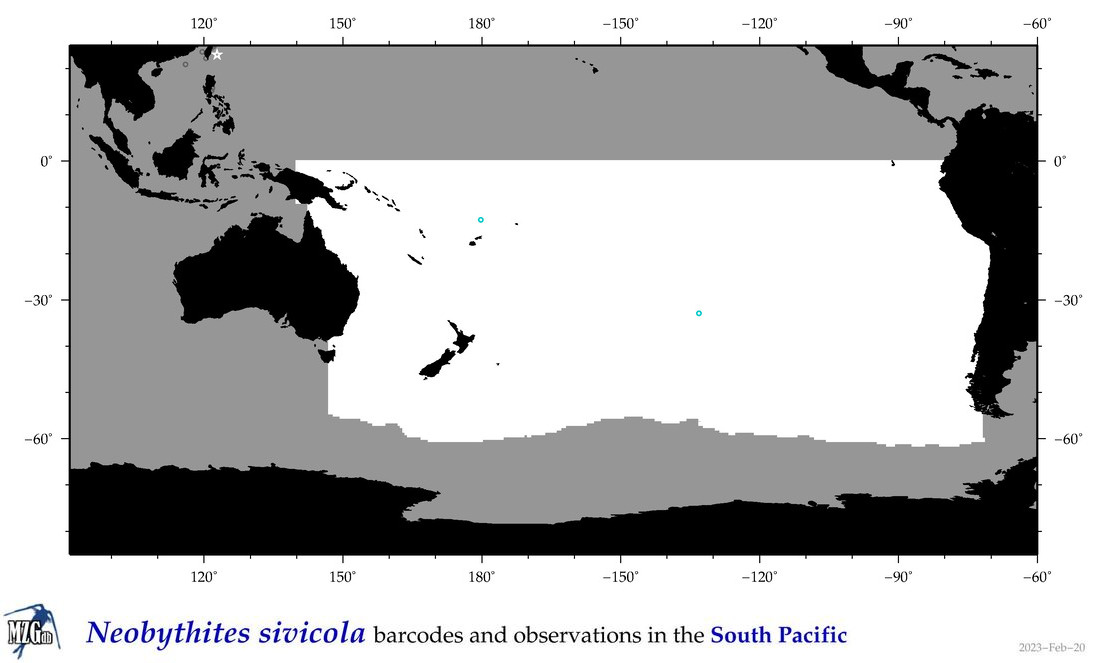 |
accepted
T5010619
R:1:0:0:0 |
| Neobythites stigmosus |
Species
(112) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 22
16S = 21
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
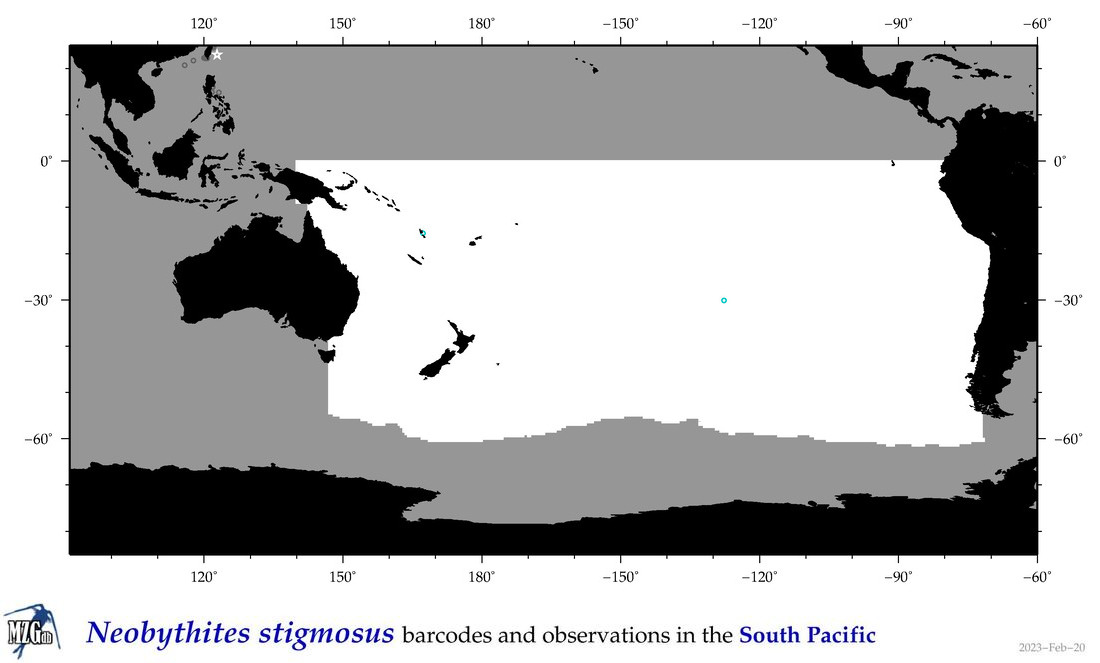 |
accepted
T5010622
R:1:0:0:0 |
| Neobythites unimaculatus |
Species
(113) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
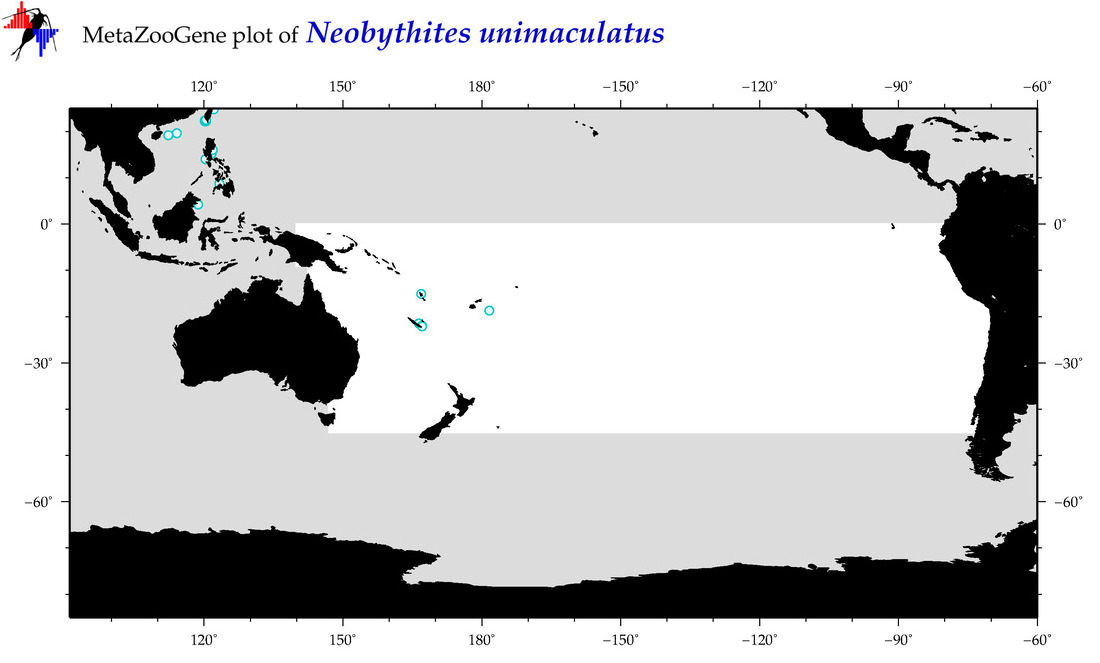 |
accepted
T5010623
R:1:0:0:0 |
| Neobythites zonatus |
Species
(114) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022589
R:1:0:0:0 |
| Ogilbia deroyi |
Species
(115) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022877
R:1:0:0:0 |
| Ogilbia ventralis |
Species
(116) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022888
R:1:0:0:0 |
| Onuxodon fowleri |
Species
(117) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 2
16S = 1
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
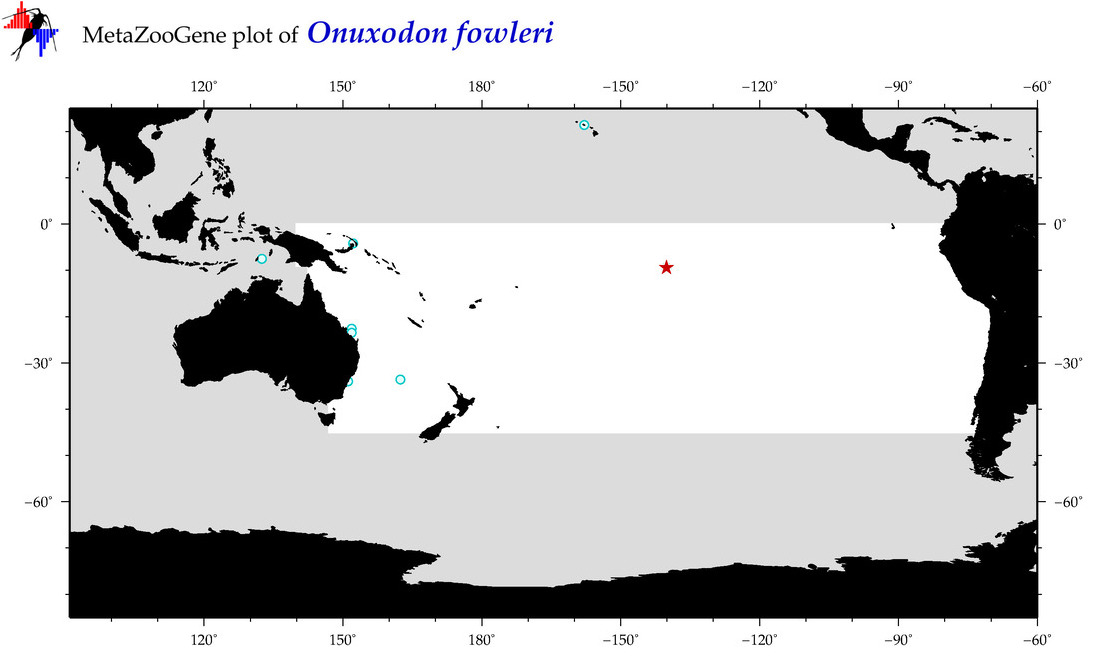 |
accepted
T5004860
R:1:0:0:0 |
| Onuxodon margaritiferae |
Species
(118) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
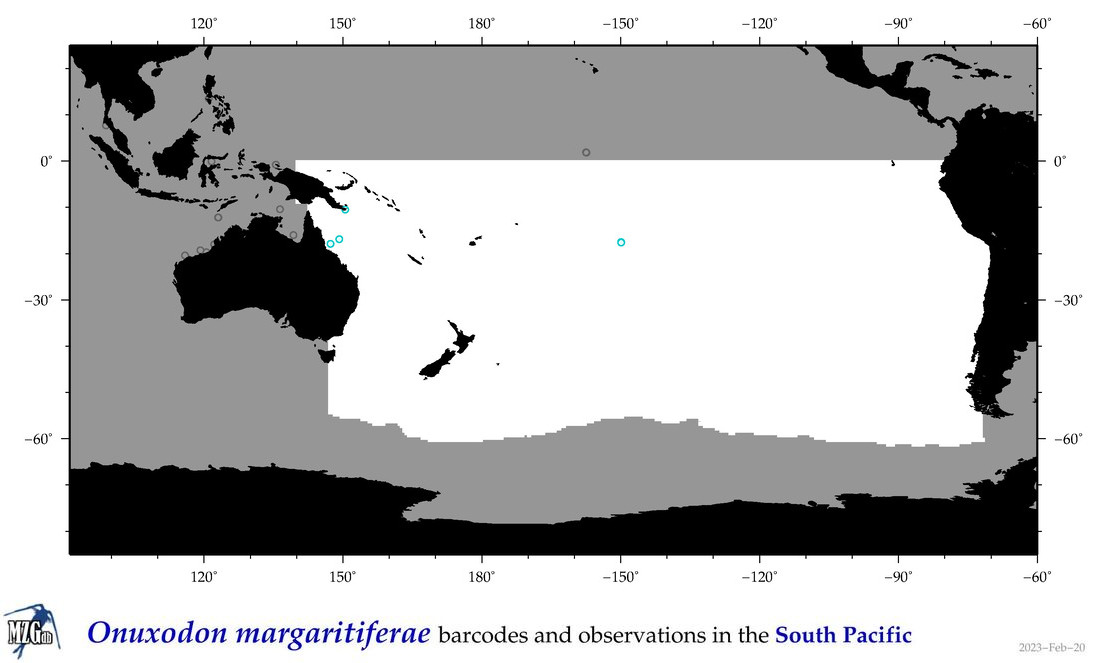 |
accepted
T5004861
R:1:0:0:0 |
| Onuxodon parvibrachium |
Species
(119) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
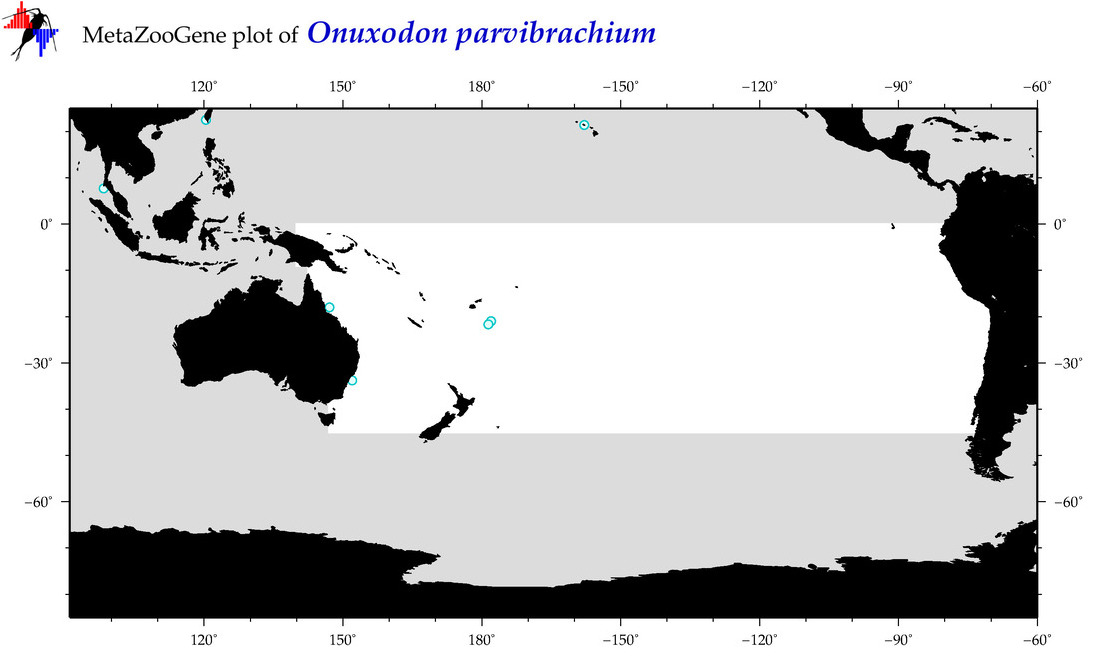 |
accepted
T5004862
R:1:0:0:0 |
| Ophidion exul |
Species
(120) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023012
R:1:0:0:0 |
| Ophidion genyopus |
Species
(121) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
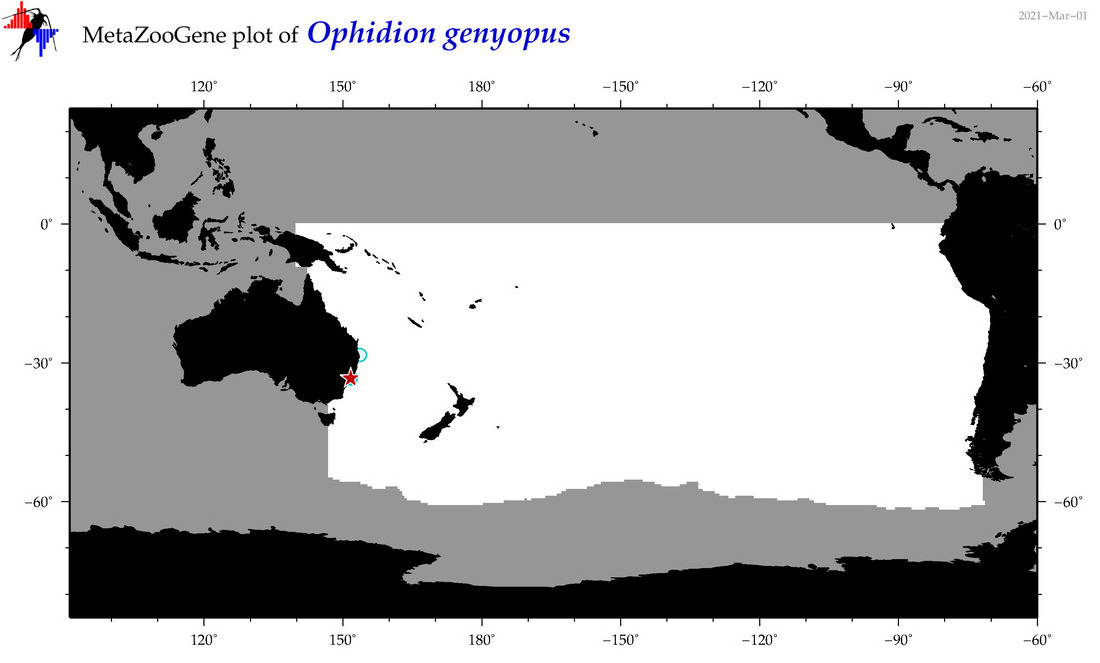 |
accepted
T5004884
R:1:0:0:0 |
| Ophidion metoecus |
Species
(122) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023021
R:1:0:0:0 |
| Ophidion muraenolepis |
Species
(123) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
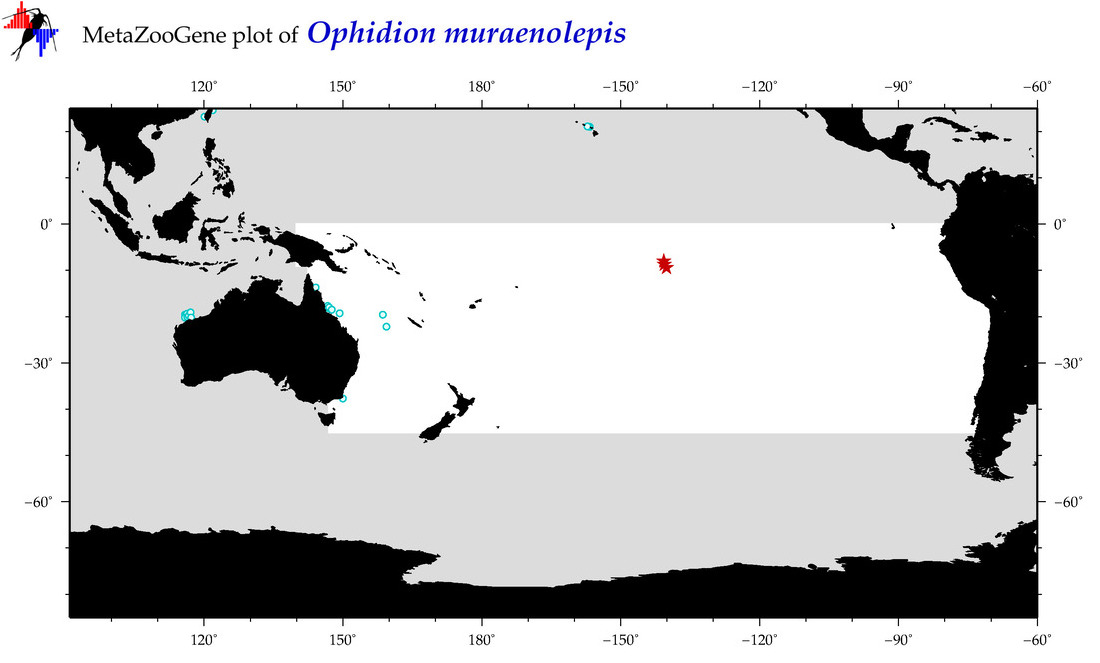 |
accepted
T5004886
R:1:0:0:0 |
| Parabrotula plagiophthalma |
Species
(124) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023361
R:1:0:0:0 |
| Paradiancistrus acutirostris |
Species
(125) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023429
R:1:0:0:0 |
| Paraphyonus bolini |
Species
(126) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
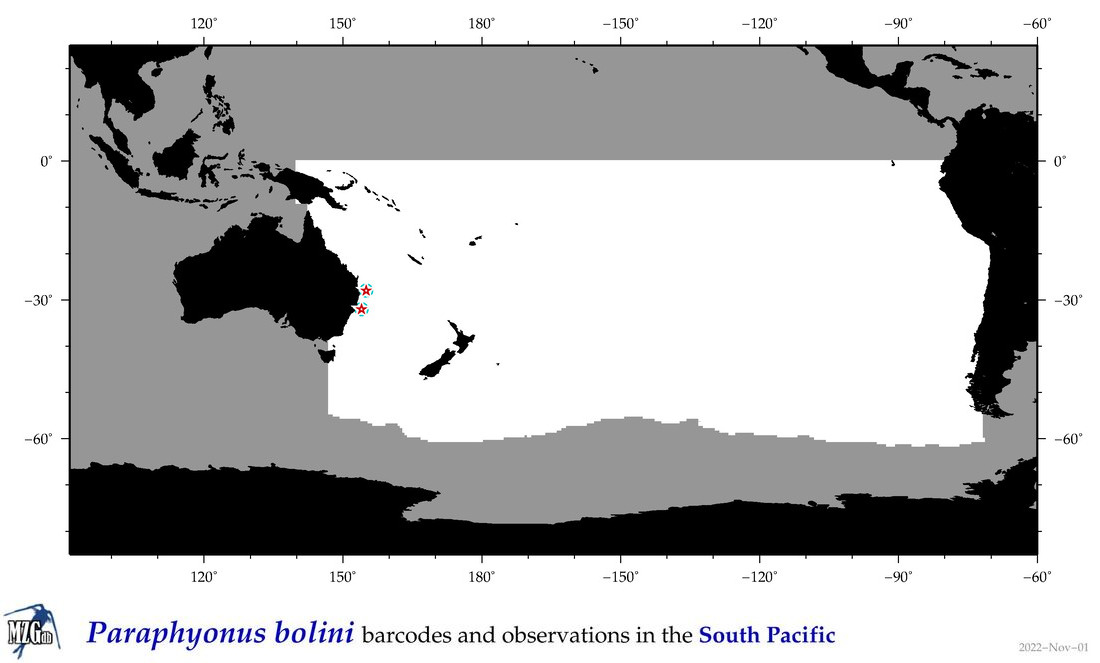 |
accepted
T5032501
R:1:0:0:0 |
| Paraphyonus rassi |
Species
(127) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
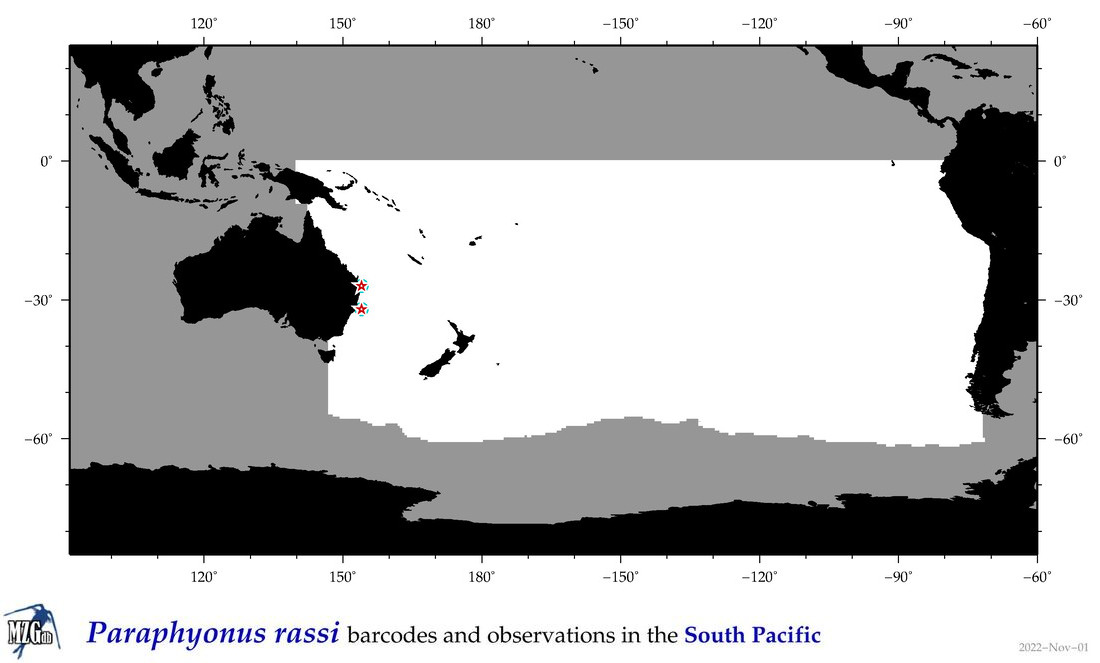 |
accepted
T5032502
R:1:0:0:0 |
| Parasaccogaster normae |
Species
(128) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028447
R:1:0:0:0 |
| Petrotyx hopkinsi |
Species
(129) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023884
R:1:0:0:0 |
| Porocephalichthys dasyrhynchus |
Species
(130) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5024362
R:1:0:0:0 |
| Porogadus melampeplus |
Species
(131) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5024366
R:1:0:0:0 |
| Pycnocraspedum armatum |
Species
(132) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5024828
R:1:0:0:0 |
| Pyramodon lindas |
Species
(133) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028578
R:1:0:0:0 |
| Pyramodon parini |
Species
(134) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028579
R:1:0:0:0 |
| Pyramodon punctatus |
Species
(135) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
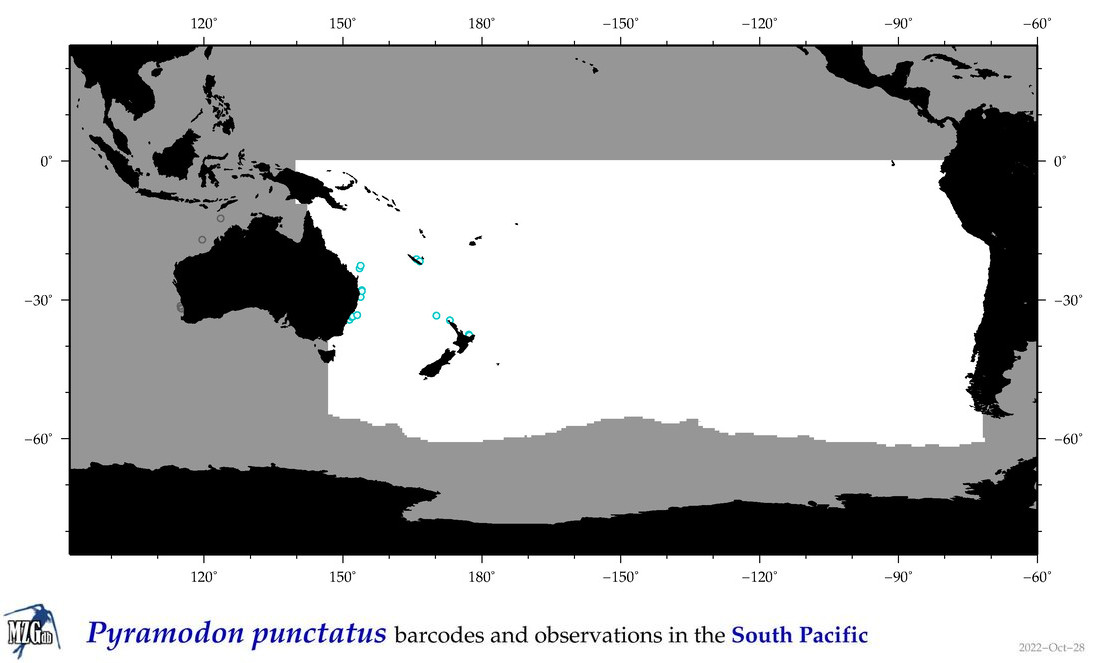 |
accepted
T5028580
R:1:0:0:0 |
| Pyramodon ventralis |
Species
(136) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
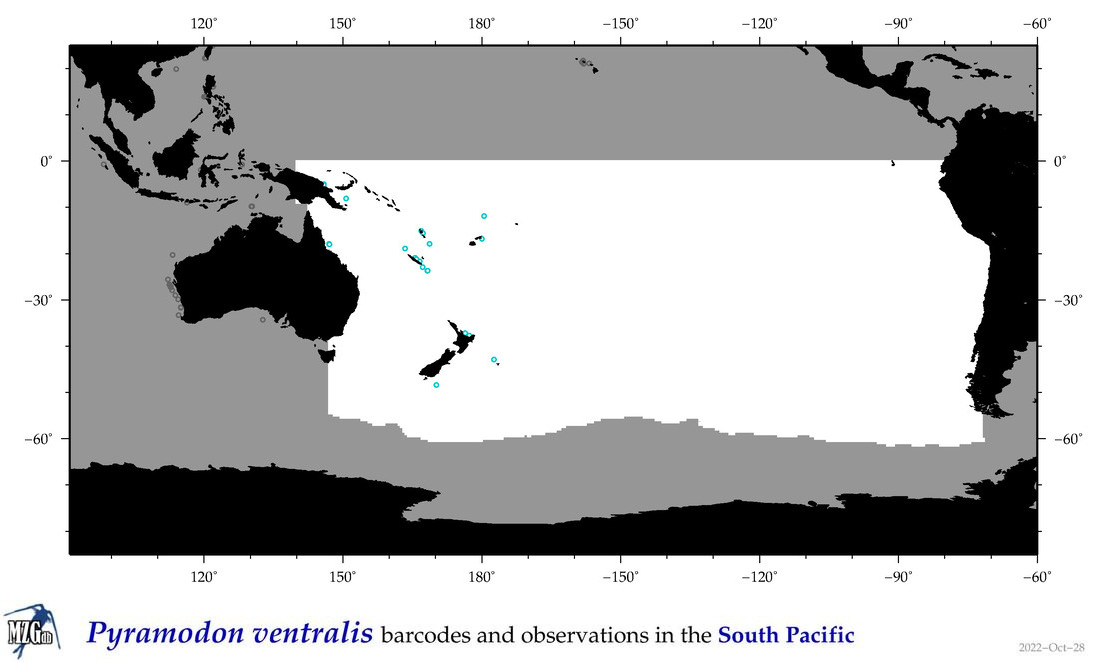 |
accepted
T5028581
R:1:0:0:0 |
| Saccogaster tuberculata |
Species
(137) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025048
R:1:0:0:0 |
| Sciadonus pedicellaris |
Species
(138) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
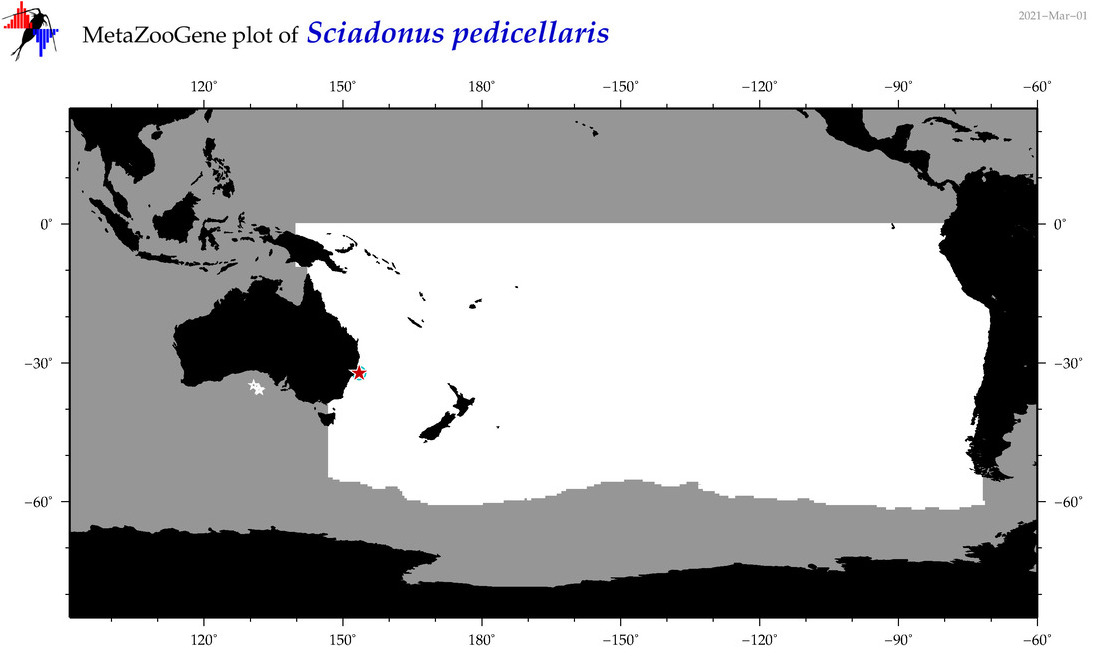 |
accepted
T5012180
R:1:0:0:0 |
| Sirembo imberbis |
Species
(139) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
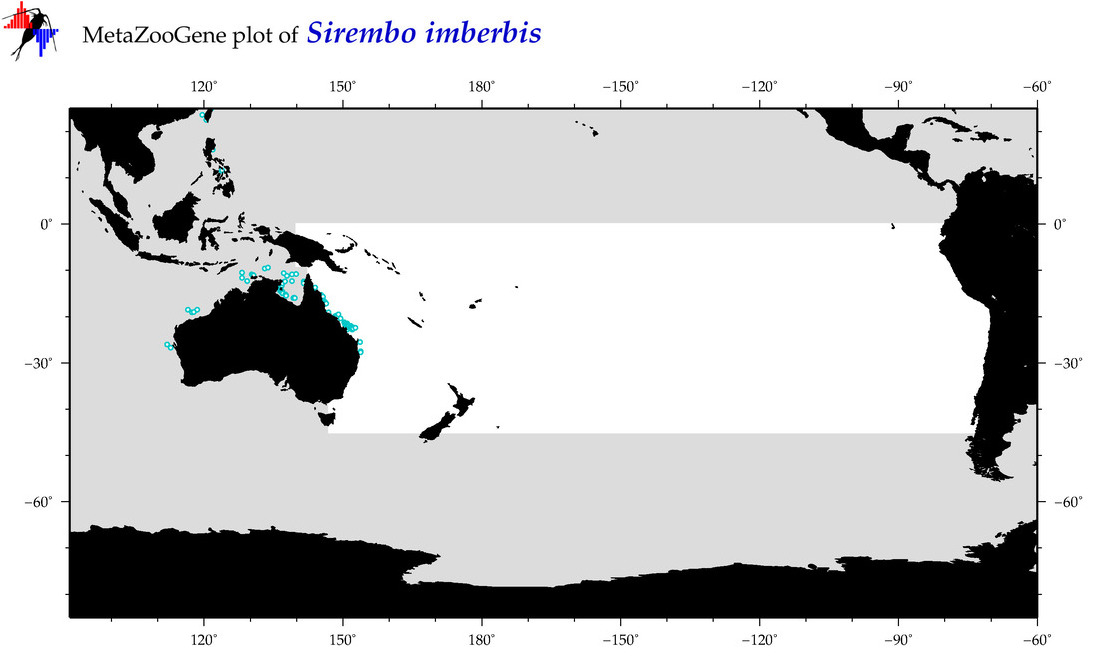 |
accepted
T5005575
R:1:0:0:0 |
| Sirembo jerdoni |
Species
(140) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
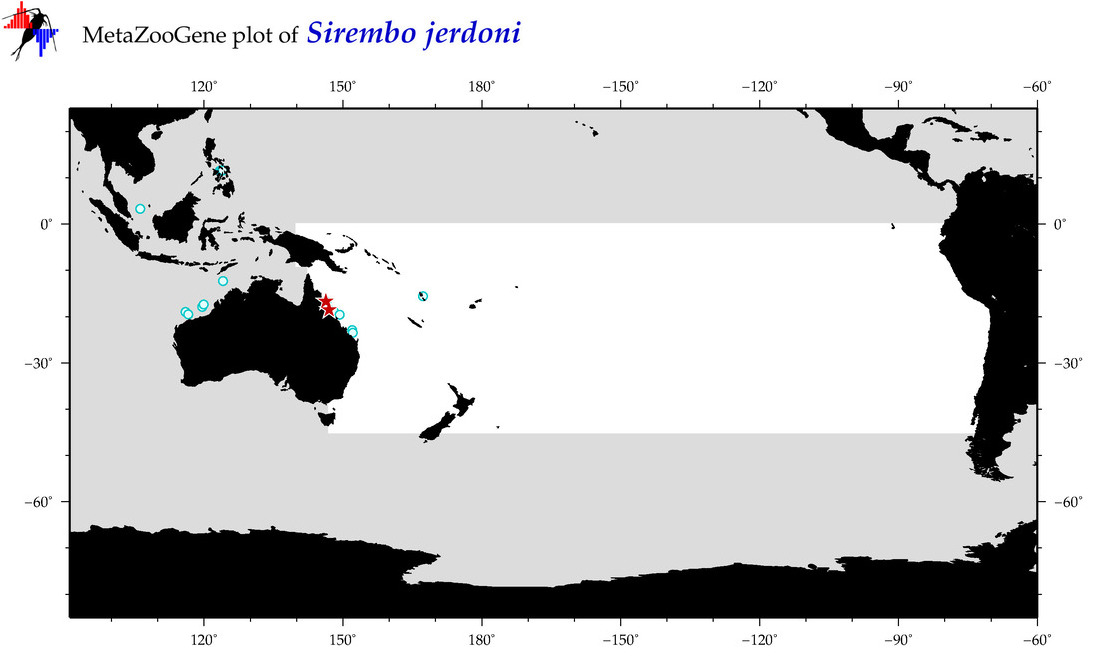 |
accepted
T5005576
R:1:0:0:0 |
| Sirembo metachroma |
Species
(141) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025605
R:1:0:0:0 |
| Snyderidia canina |
Species
(142) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025614
R:1:0:0:0 |
| Spectrunculus grandis |
Species
(143) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
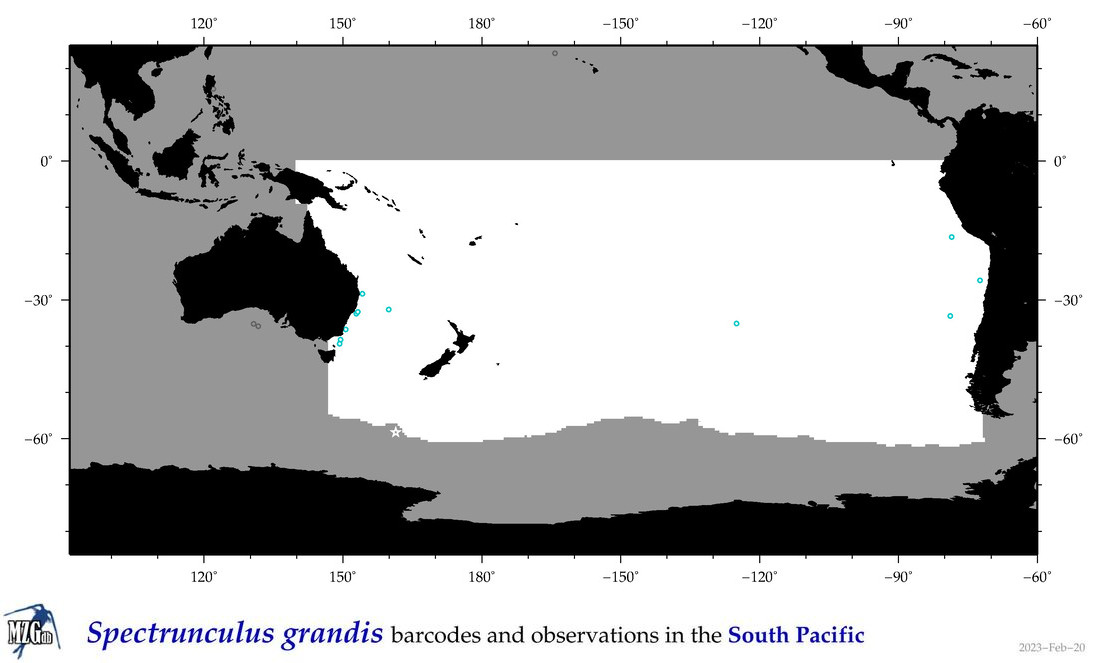 |
accepted
T5001271
R:1:0:0:0 |
| Spottobrotula amaculata |
Species
(144) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025709
R:1:0:0:0 |
| Tauredophidium hextii |
Species
(145) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026137
R:1:0:0:0 |
| Thalassobathia nelsoni |
Species
(146) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028727
R:1:0:0:0 |
| Thermichthys hollisi |
Species
(147) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026202
R:1:0:0:0 |
| Typhlonus nasus |
Species
(148) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026496
R:1:0:0:0 |
| Ventichthys biospeedoi |
Species
(149) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026608
R:1:0:0:0 |