Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2023-m07-15
2023-Jul-20 |
| Ahliesaurus brevis |
Species
(1) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001987
R:1:0:0:0 |
| Alepisaurus brevirostris |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003059
R:1:0:0:0 |
| Alepisaurus ferox |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 29
12S = 8
16S = 4
18S = 1
28S = 0
|
COI = 4
12S = 2
16S = 2
18S = 0
28S = 0
|
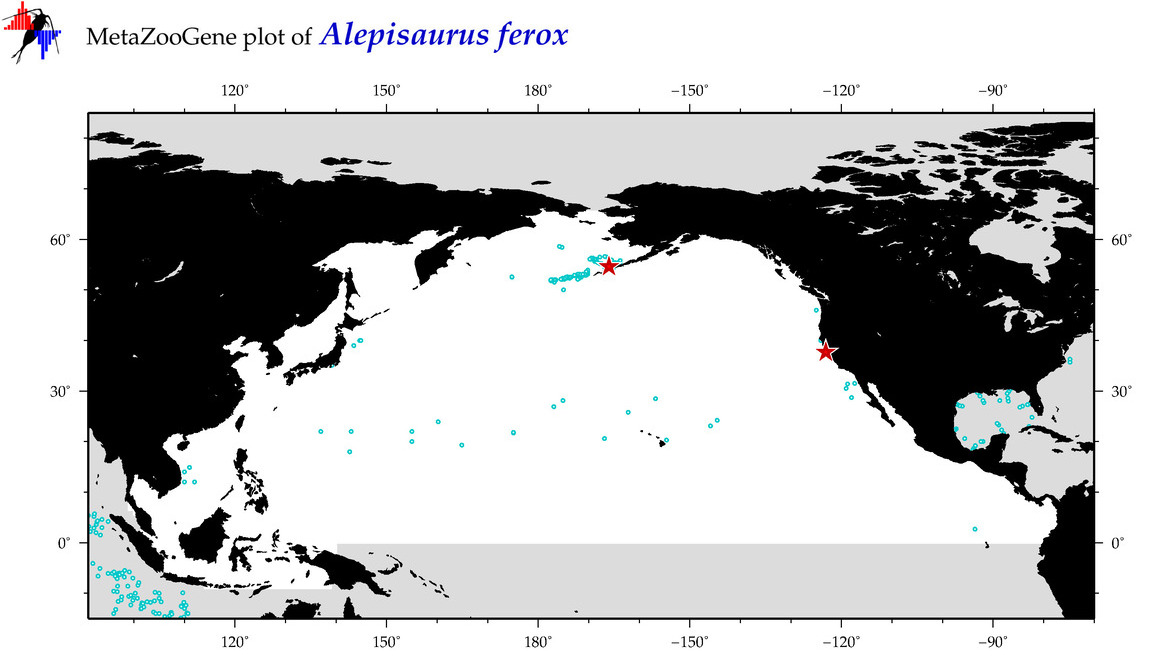 |
accepted
T5000142
R:1:0:0:0 |
| Anotopterus nikparini |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003118
R:1:0:0:0 |
| Anotopterus pharao |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000174
R:1:0:0:0 |
| Arctozenus risso |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 25
12S = 3
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
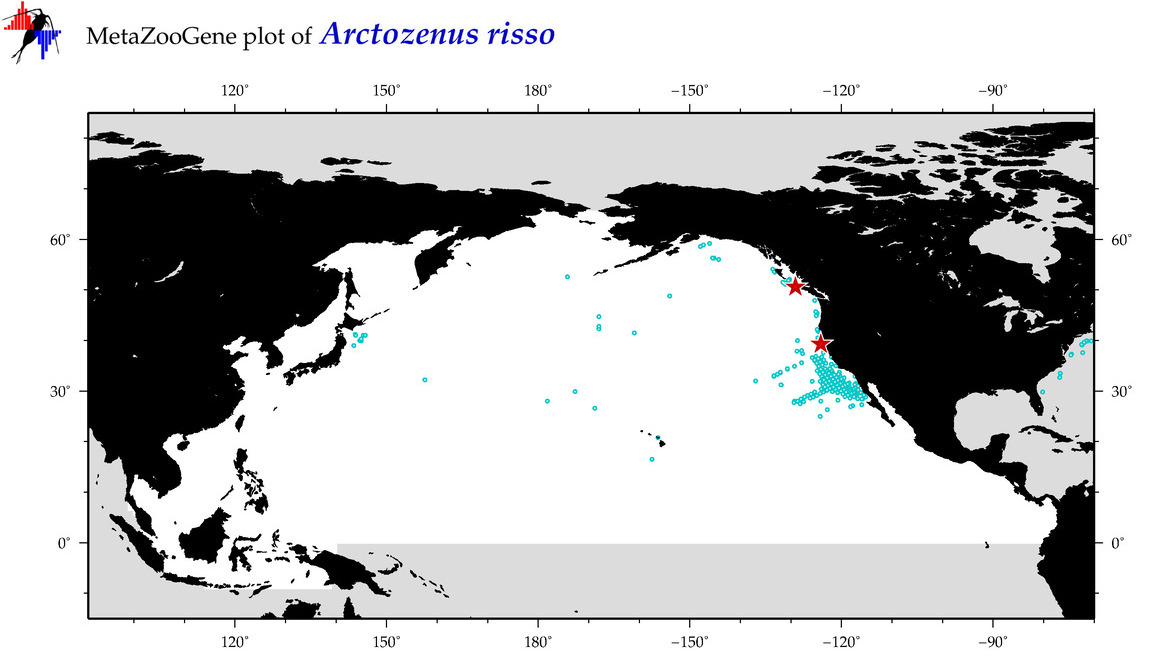 |
accepted
T5000192
R:1:0:0:0 |
| Aulopus bajacali |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001996
R:1:0:0:0 |
| Bathymicrops regis |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003248
R:1:0:0:0 |
| Bathypterois atricolor |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
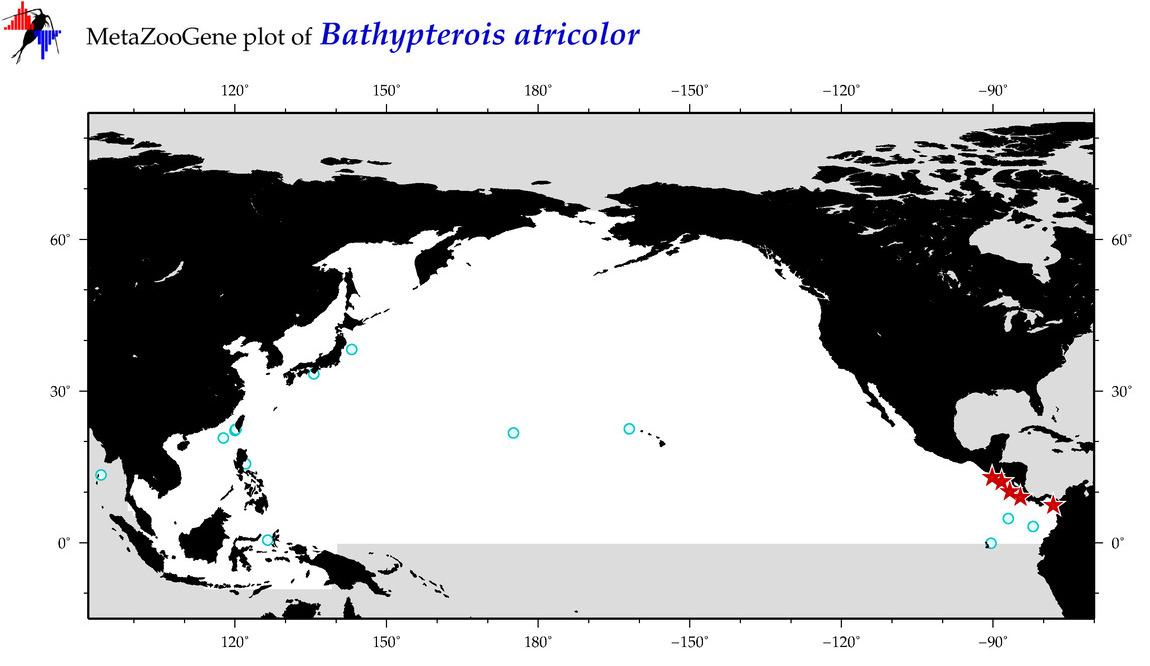 |
accepted
T5003251
R:1:0:0:0 |
| Bathypterois grallator |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003253
R:1:0:0:0 |
| Bathypterois guentheri |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
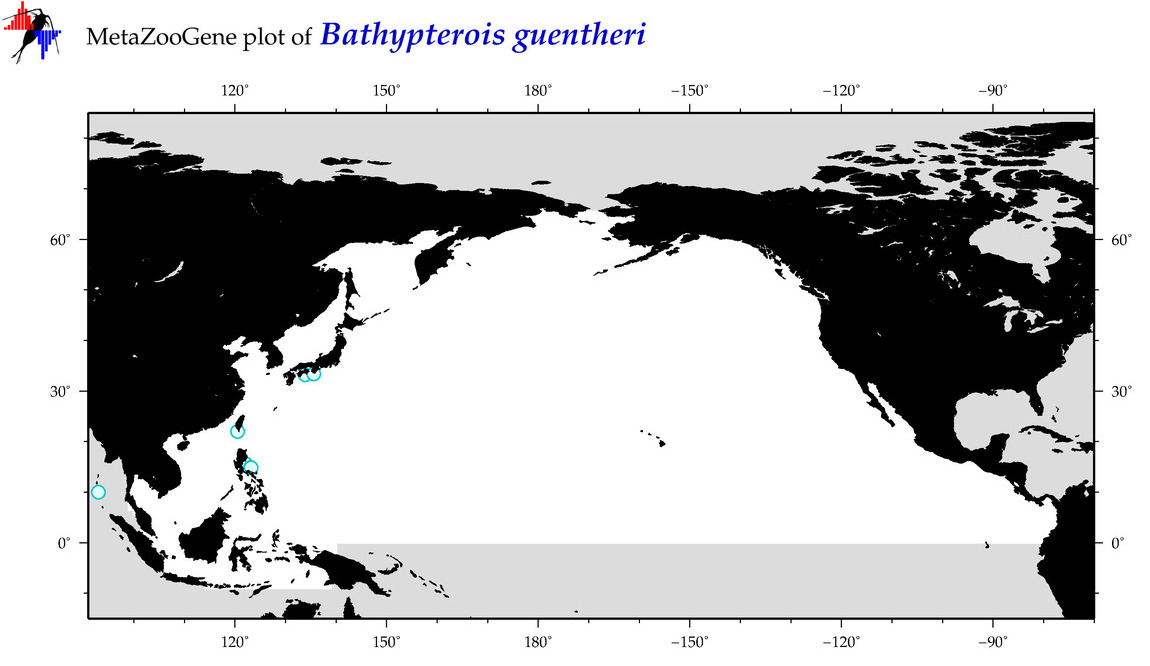 |
accepted
T5003254
R:1:0:0:0 |
| Bathypterois longipes |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017062
R:1:0:0:0 |
| Bathypterois ventralis |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 11
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003258
R:1:0:0:0 |
| Bathysauroides gigas |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017080
R:1:0:0:0 |
| Bathysaurus mollis |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5017082
R:1:0:0:0 |
| Bathytyphlops marionae |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
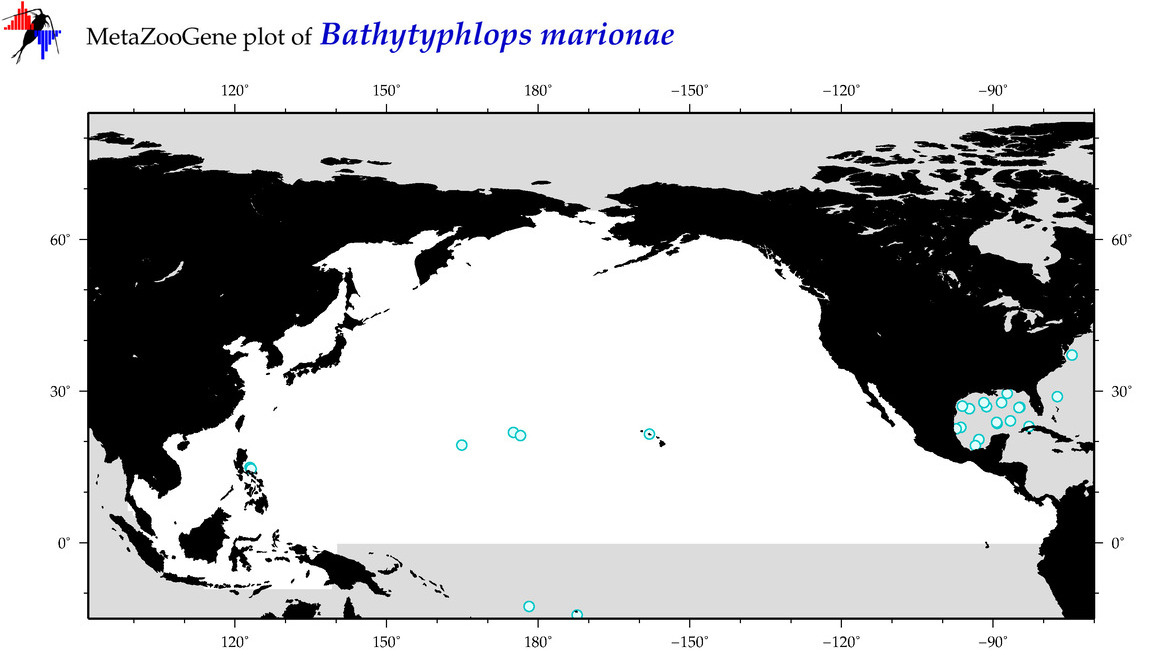 |
accepted
T5000272
R:1:0:0:0 |
| Benthalbella dentata |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 15
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000277
R:1:0:0:0 |
| Benthalbella infans |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
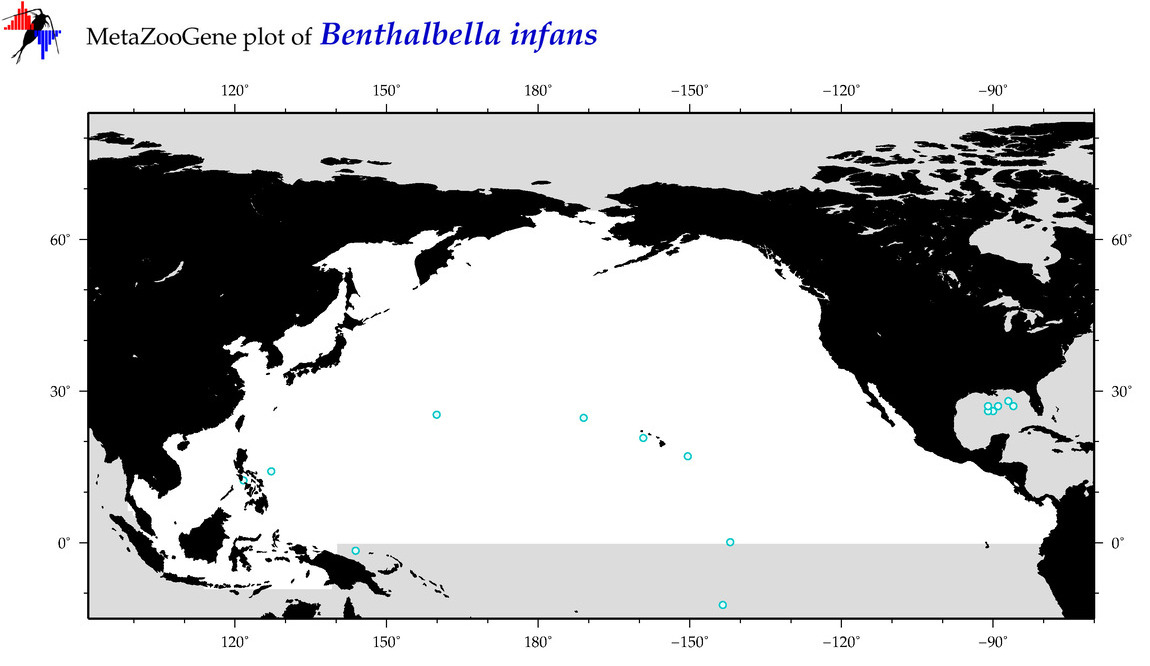 |
accepted
T5000278
R:1:0:0:0 |
| Benthalbella linguidens |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 4
16S = 2
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
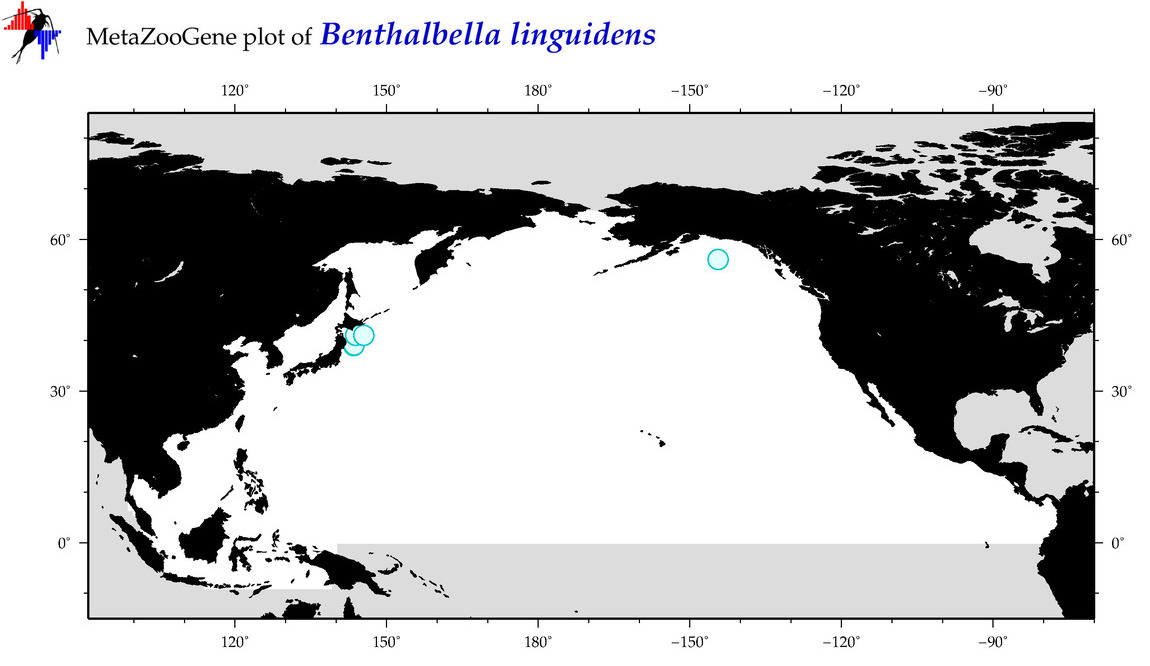 |
accepted
T5003276
R:1:0:0:0 |
| Chlorophthalmus acutifrons |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 26
12S = 3
16S = 2
18S = 0
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003527
R:1:0:0:0 |
| Chlorophthalmus agassizi |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 37
12S = 1
16S = 2
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000402
R:1:1:0:0 |
| Chlorophthalmus albatrossis |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 6
16S = 5
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
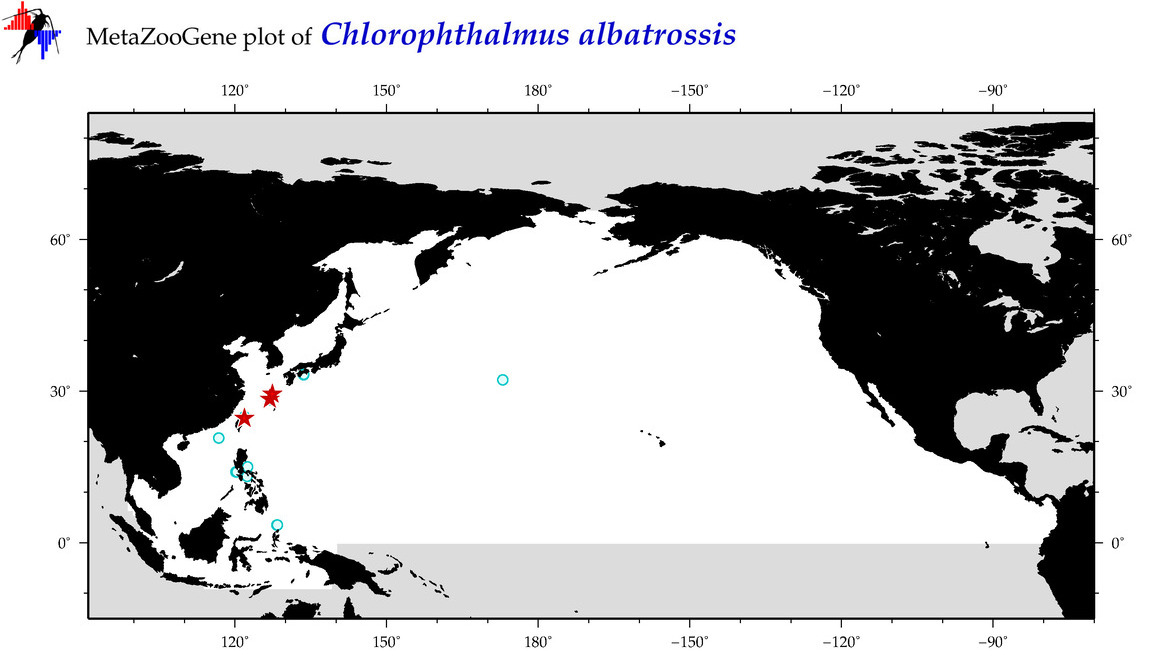 |
accepted
T5003528
R:1:0:0:0 |
| Chlorophthalmus borealis |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 3
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
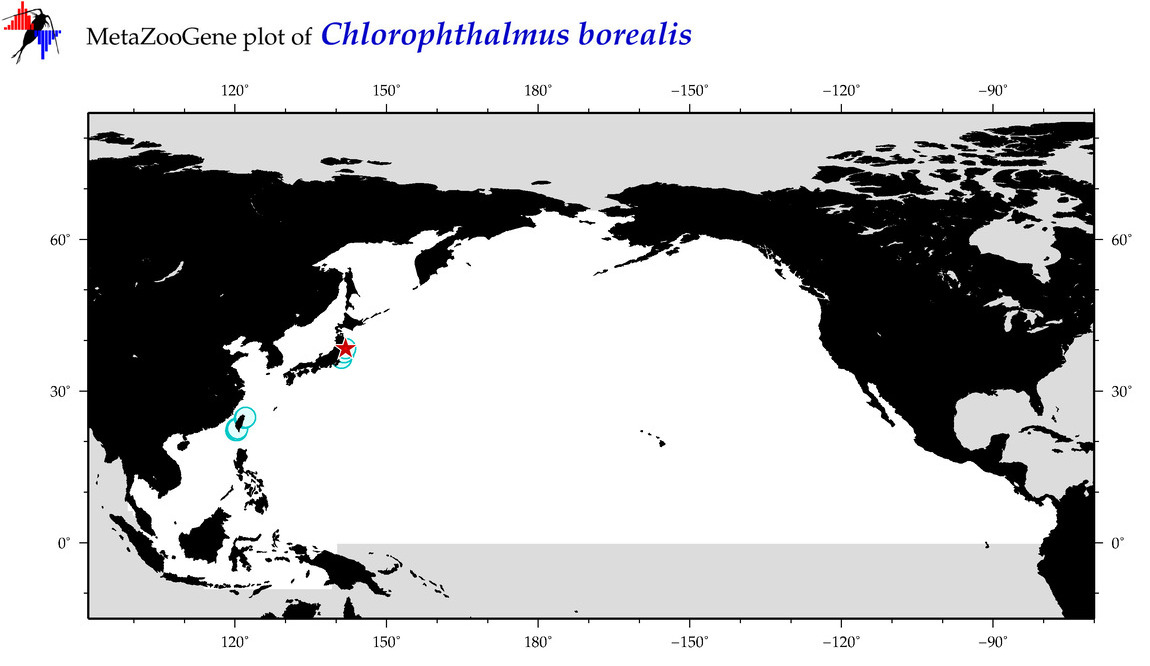 |
accepted
T5003529
R:1:0:0:0 |
| Chlorophthalmus mento |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
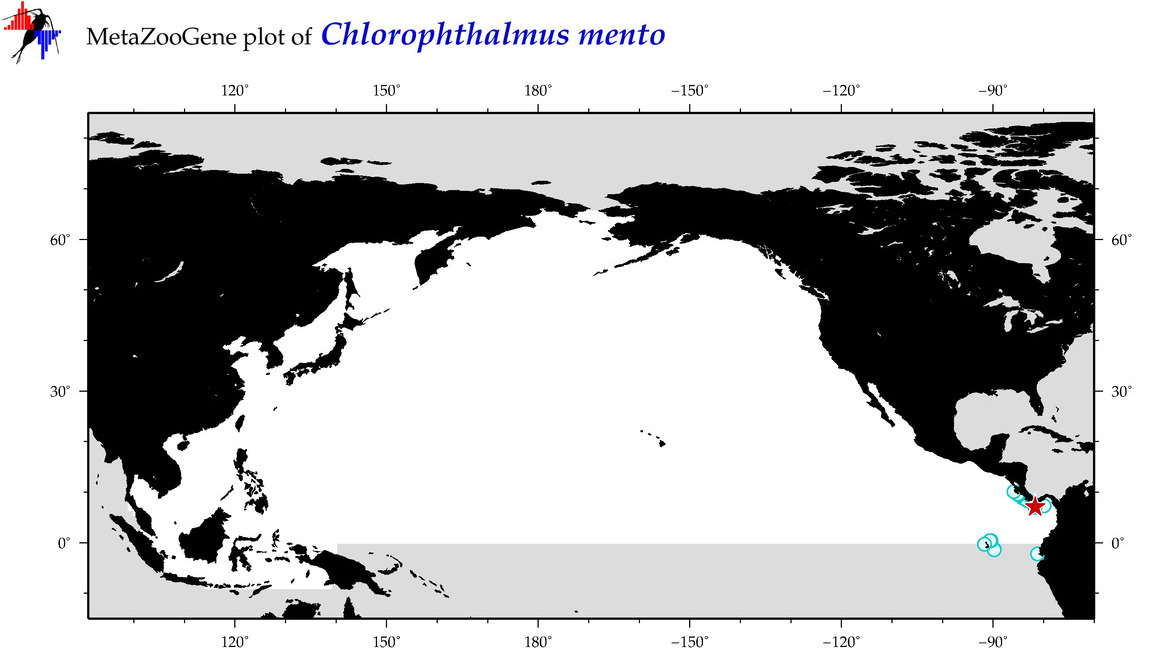 |
accepted
T5003531
R:1:0:0:0 |
| Chlorophthalmus nigromarginatus |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 4
16S = 3
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
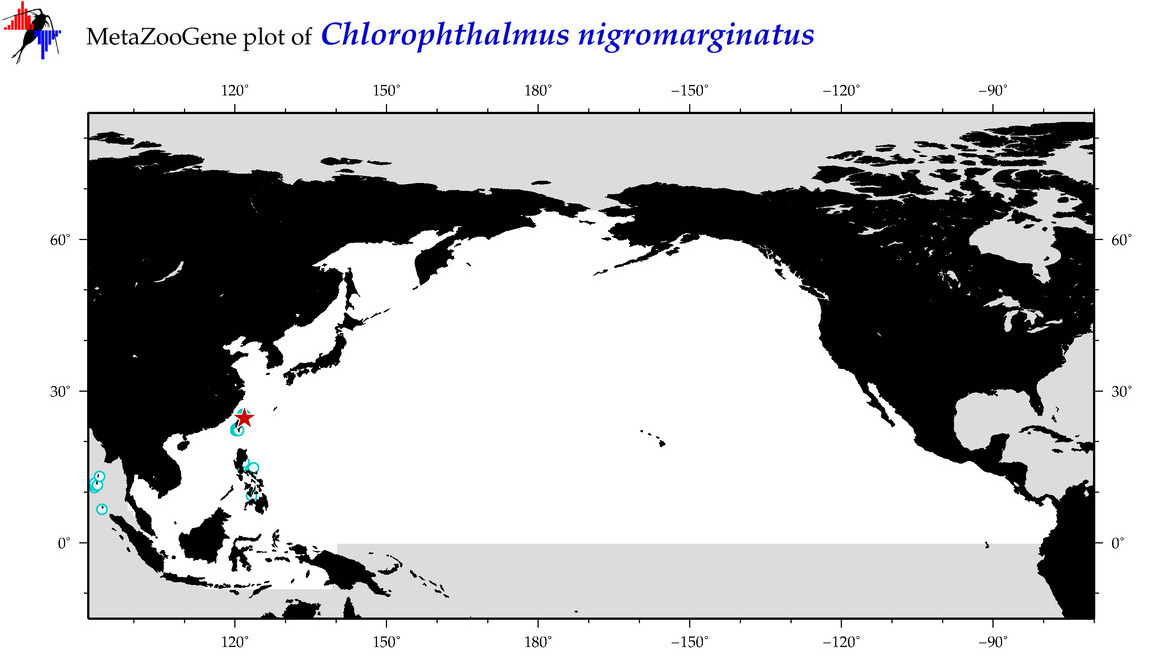 |
accepted
T5003532
R:1:0:0:0 |
| Chlorophthalmus pectoralis |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003533
R:1:0:0:0 |
| Chlorophthalmus proridens |
Species
(27) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018069
R:1:0:0:0 |
| Coccorella atlantica |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 4
16S = 2
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001827
R:1:0:0:0 |
| Coccorella atrata |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 3
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018422
R:1:0:0:0 |
| Evermannella ahlstromi |
Species
(30) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001990
R:1:0:0:0 |
| Evermannella indica |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
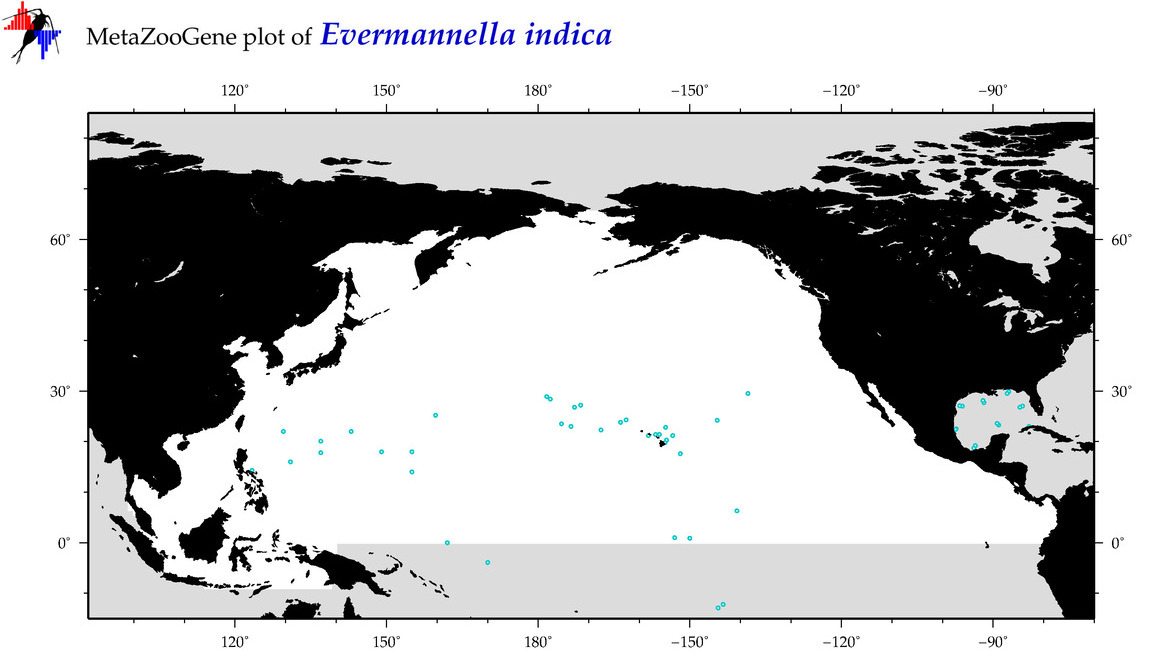 |
accepted
T5001828
R:1:0:0:0 |
| Gigantura chuni |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004059
R:1:0:0:0 |
| Gigantura indica |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
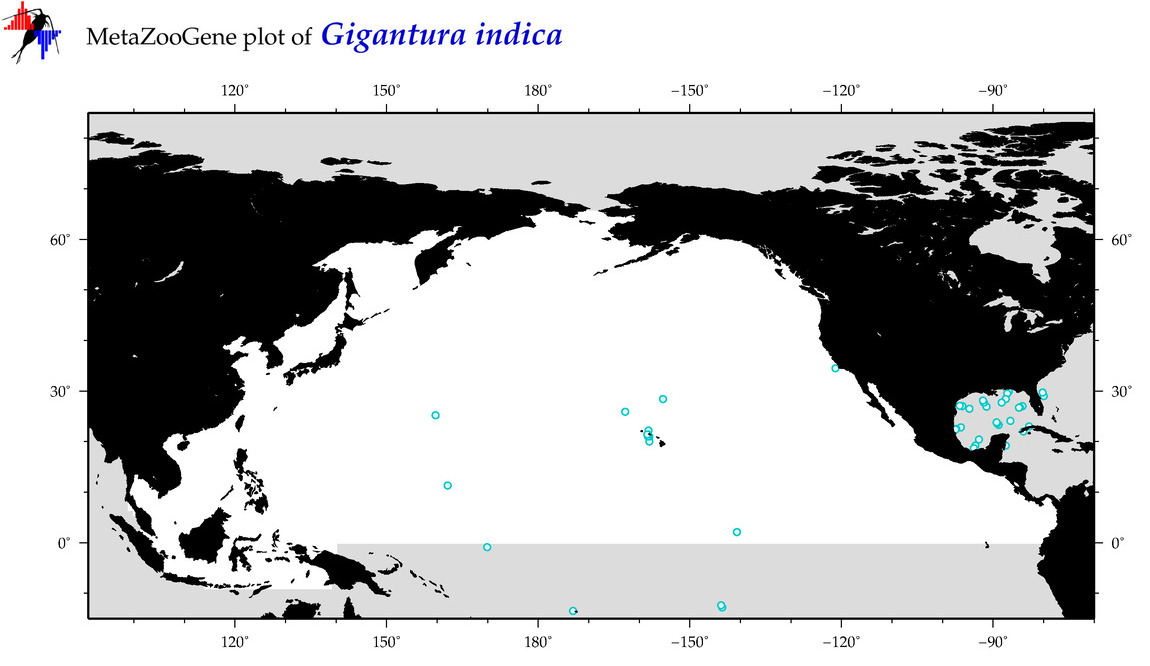 |
accepted
T5000598
R:1:0:0:0 |
| Harpadon microchir |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 3
16S = 2
18S = 0
28S = 1
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5009541
R:1:0:0:0 |
| Harpadon nehereus |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 132
12S = 5
16S = 14
18S = 11
28S = 0
|
COI = 32
12S = 0
16S = 11
18S = 11
28S = 0
|
 |
accepted
T5009542
R:1:1:0:0 |
| Harpadon translucens |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020620
R:1:1:0:0 |
| Hime damasi |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
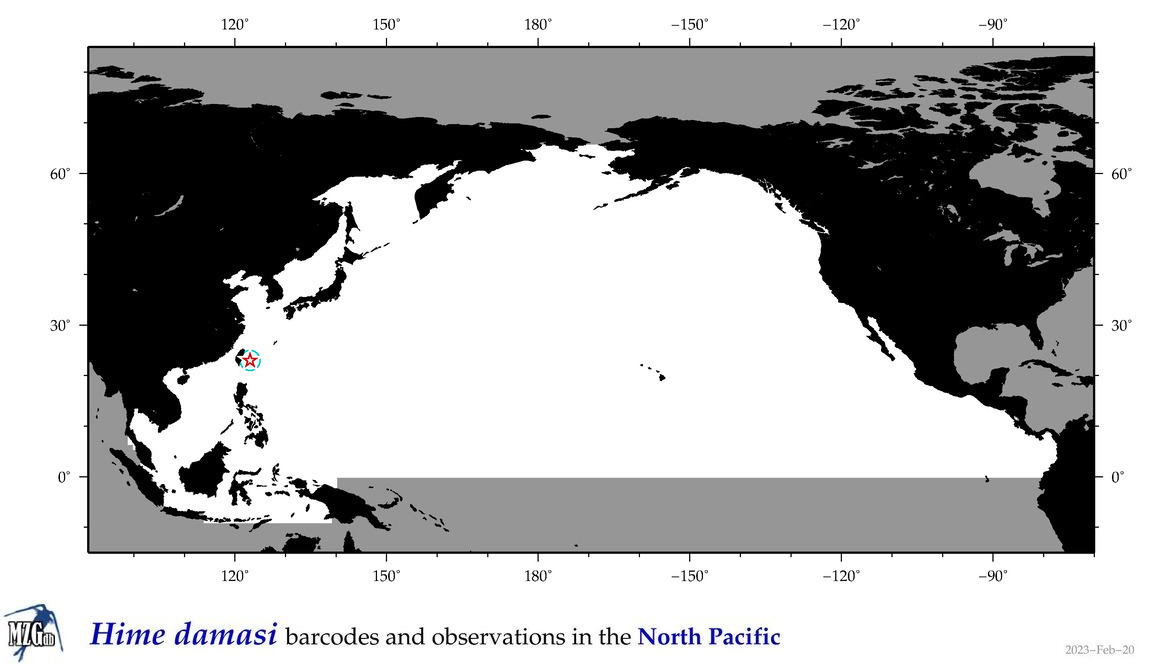 |
accepted
T5004242
R:1:0:0:0 |
| Hime formosana |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5020848
R:1:0:0:0 |
| Hime japonica |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 4
16S = 3
18S = 0
28S = 1
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
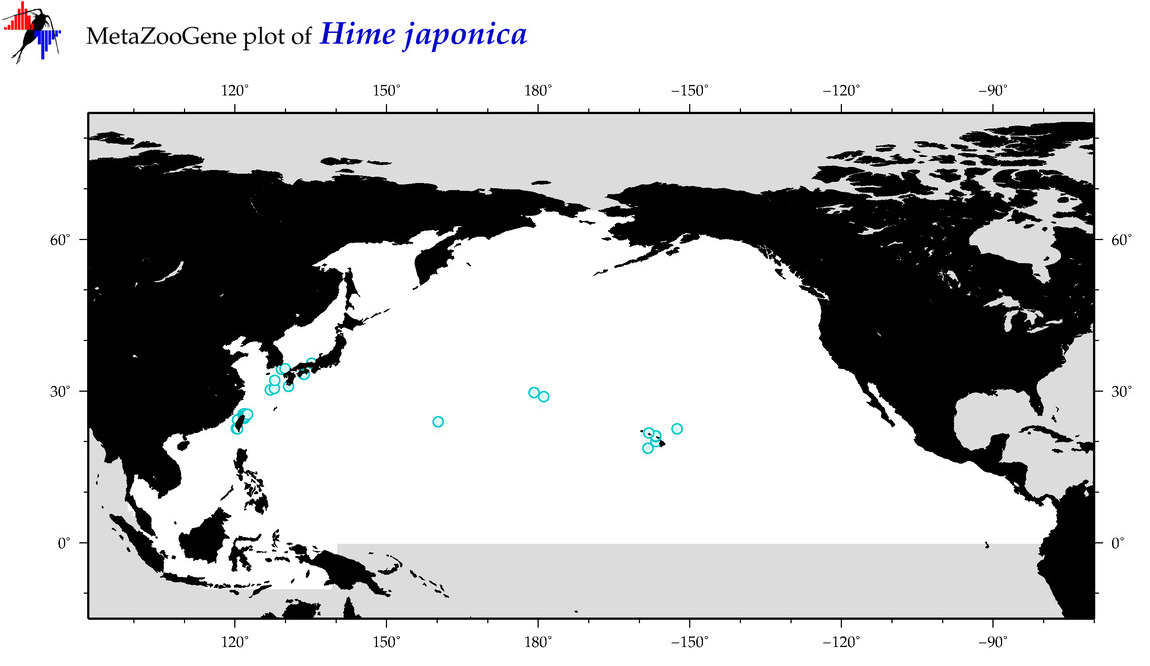 |
accepted
T5004244
R:1:0:0:0 |
| Hime pyrhistion |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
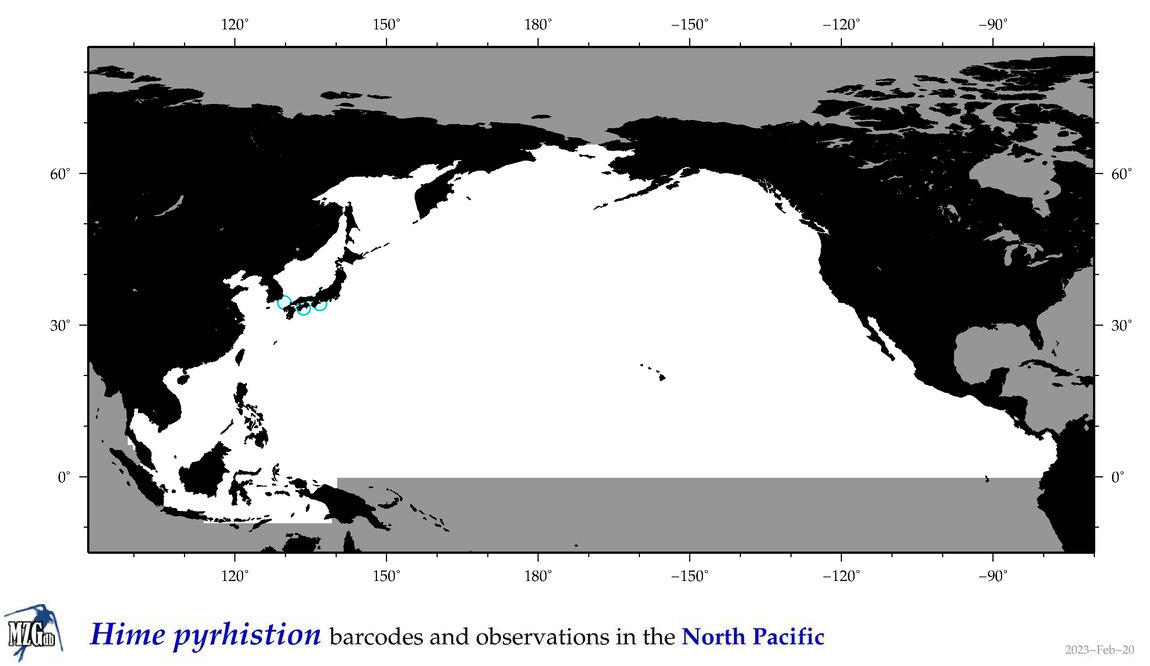 |
accepted
T5004245
R:1:0:0:0 |
| Hime surrubea |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028117
R:1:0:0:0 |
| Ipnops agassizii |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021138
R:1:0:0:0 |
| Ipnops meadi |
Species
(43) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021139
R:1:0:0:0 |
| Leptaulopus damasi |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028215
R:1:0:0:0 |
| Lestidiops bathyopteryx |
Species
(45) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028216
R:1:0:0:0 |
| Lestidiops distans |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021565
R:1:0:0:0 |
| Lestidiops jayakari |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000790
R:1:0:0:0 |
| Lestidiops mirabilis |
Species
(48) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021567
R:1:0:0:0 |
| Lestidiops neles |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
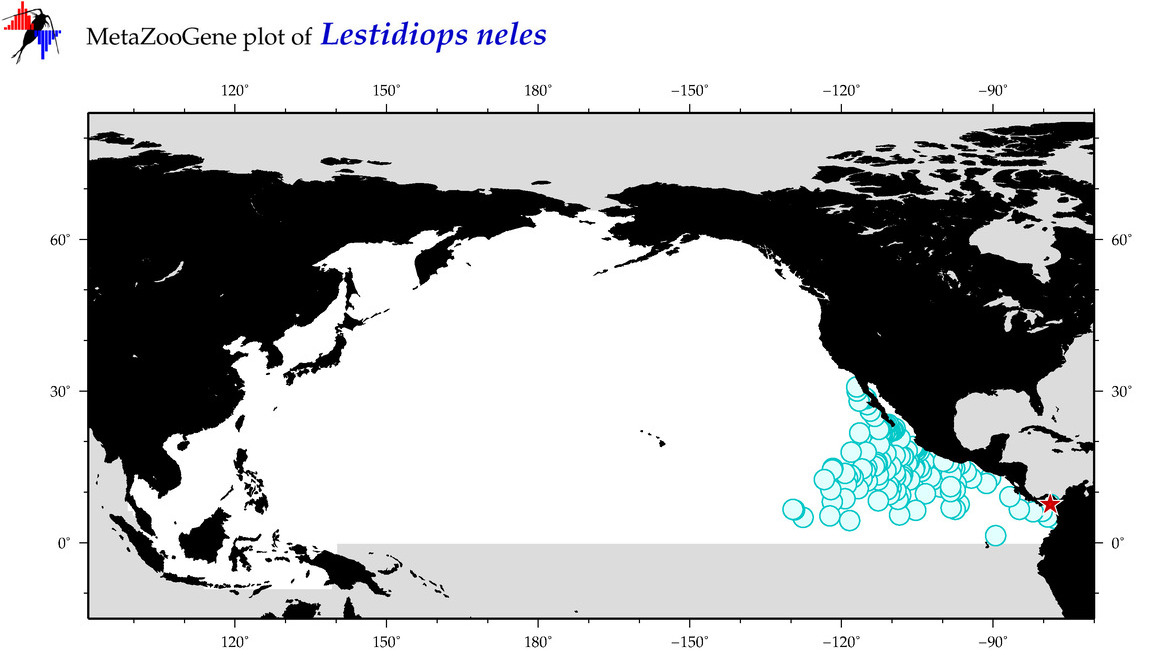 |
accepted
T5001991
R:1:0:0:0 |
| Lestidiops pacificus |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
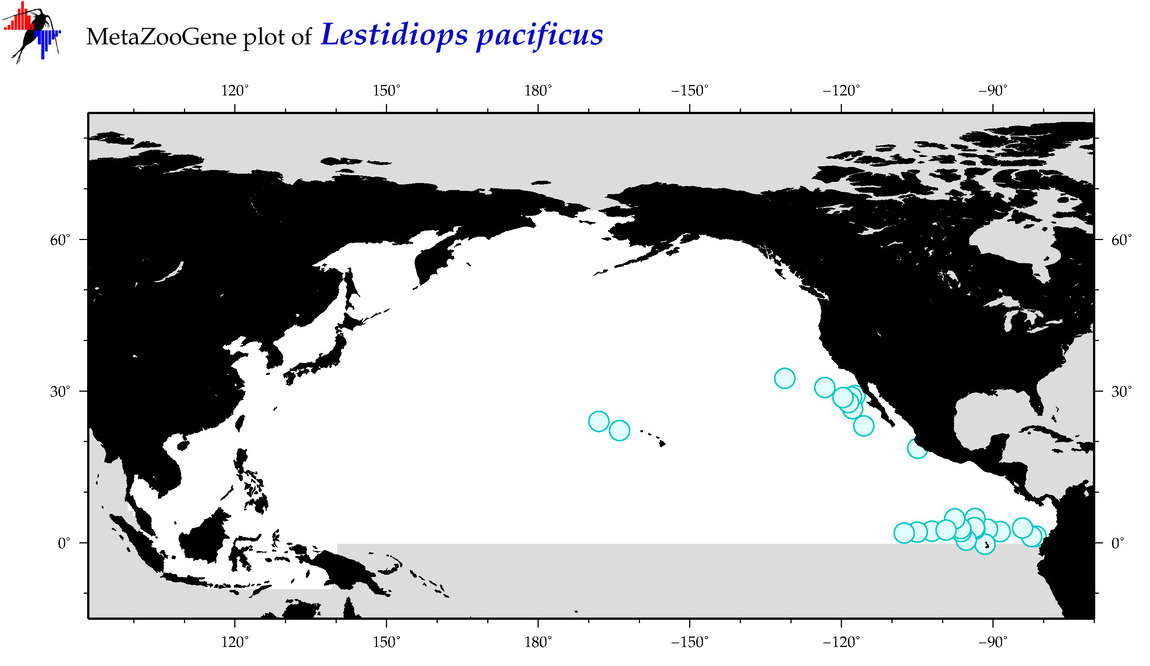 |
accepted
T5001992
R:1:0:0:0 |
| Lestidiops ringens |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 3
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
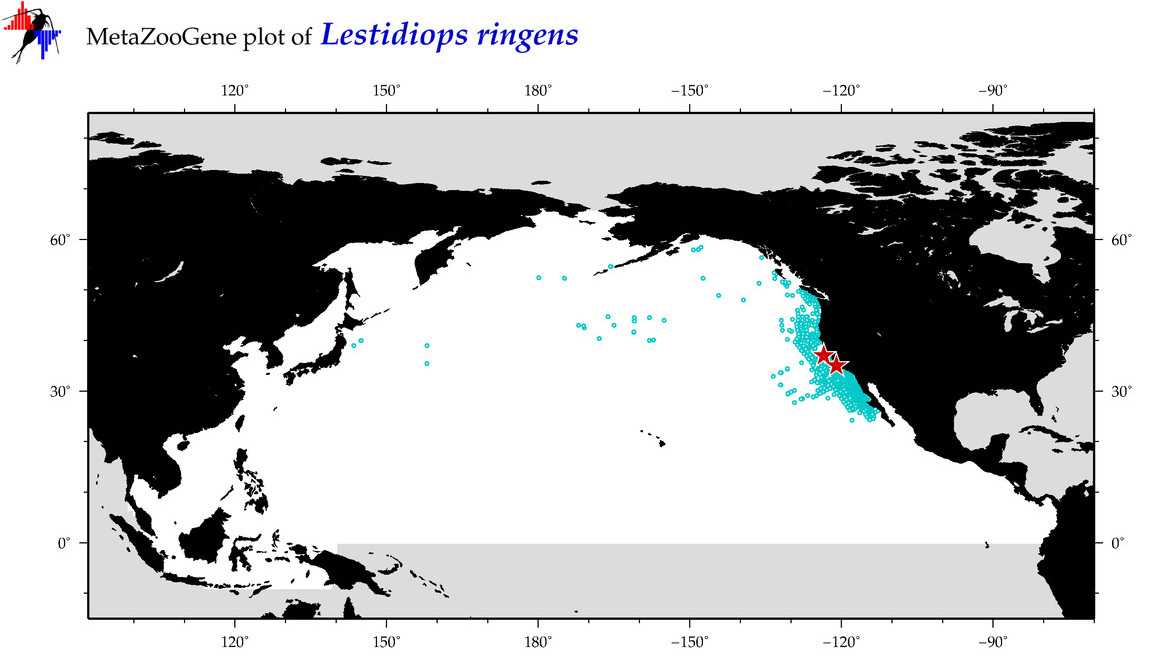 |
accepted
T5000038
R:1:0:0:0 |
| Lestidiops sphyraenopsis |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021569
R:1:0:0:0 |
| Lestidium atlanticum |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
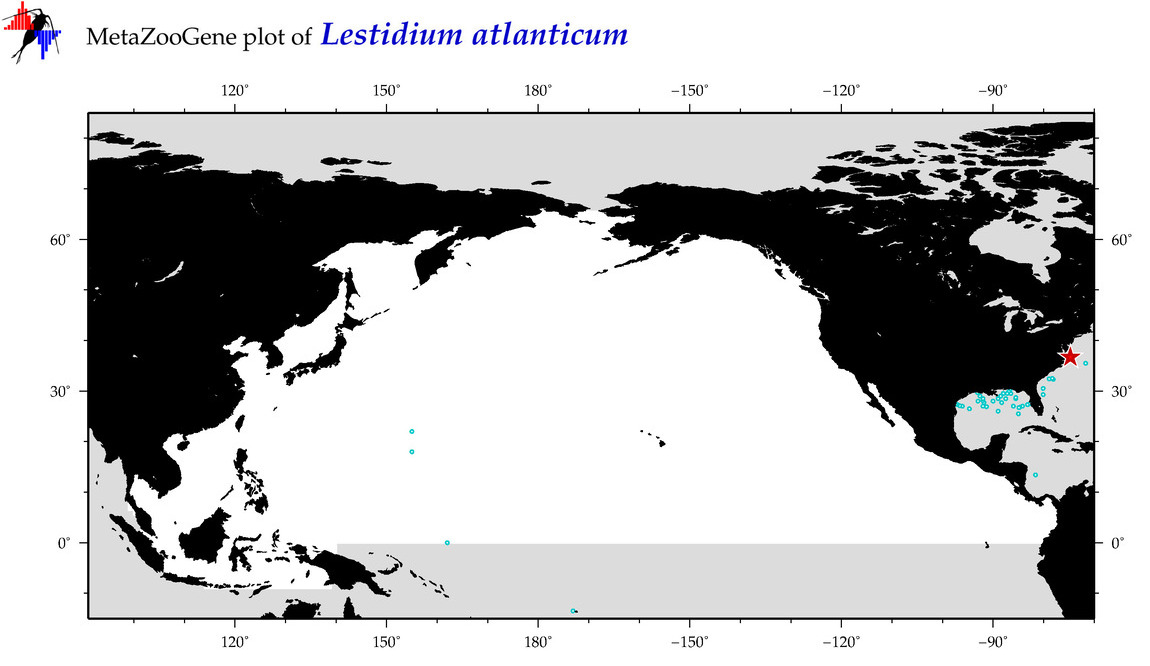 |
accepted
T5000792
R:1:0:0:0 |
| Lestidium bigelowi |
Species
(54) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028217
R:1:0:0:0 |
| Lestidium nudum |
Species
(55) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021571
R:1:0:0:0 |
| Lestidium prolixum |
Species
(56) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5013117
R:1:0:0:0 |
| Lestrolepis intermedia |
Species
(57) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
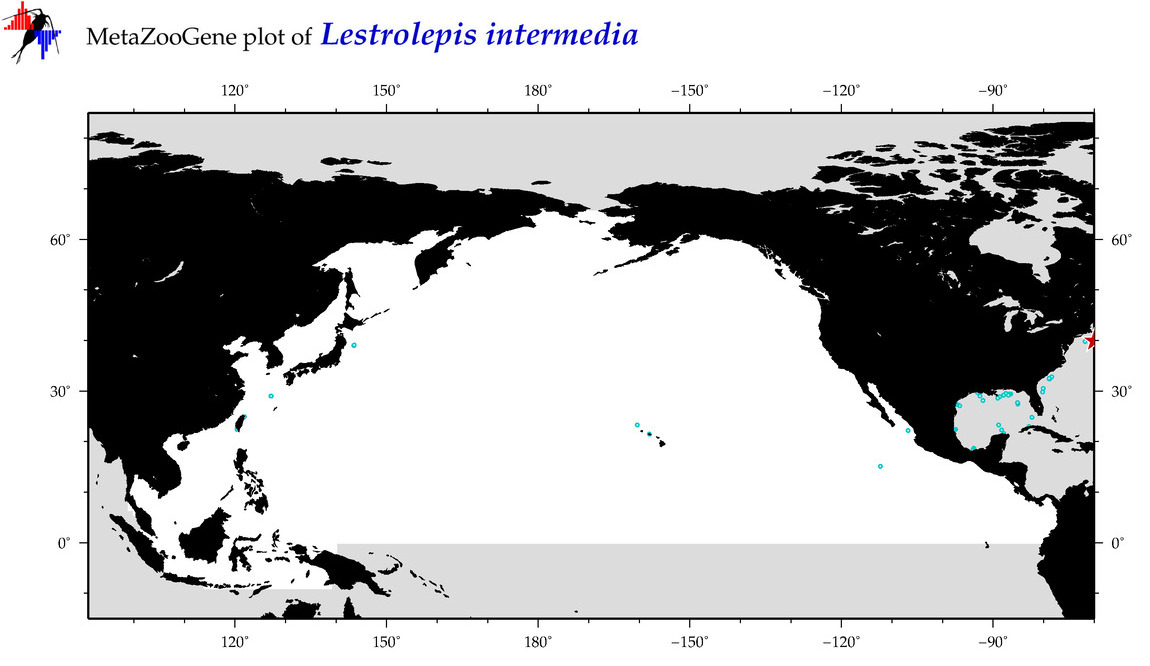 |
accepted
T5000793
R:1:0:0:0 |
| Lestrolepis japonica |
Species
(58) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 1
16S = 1
18S = 0
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
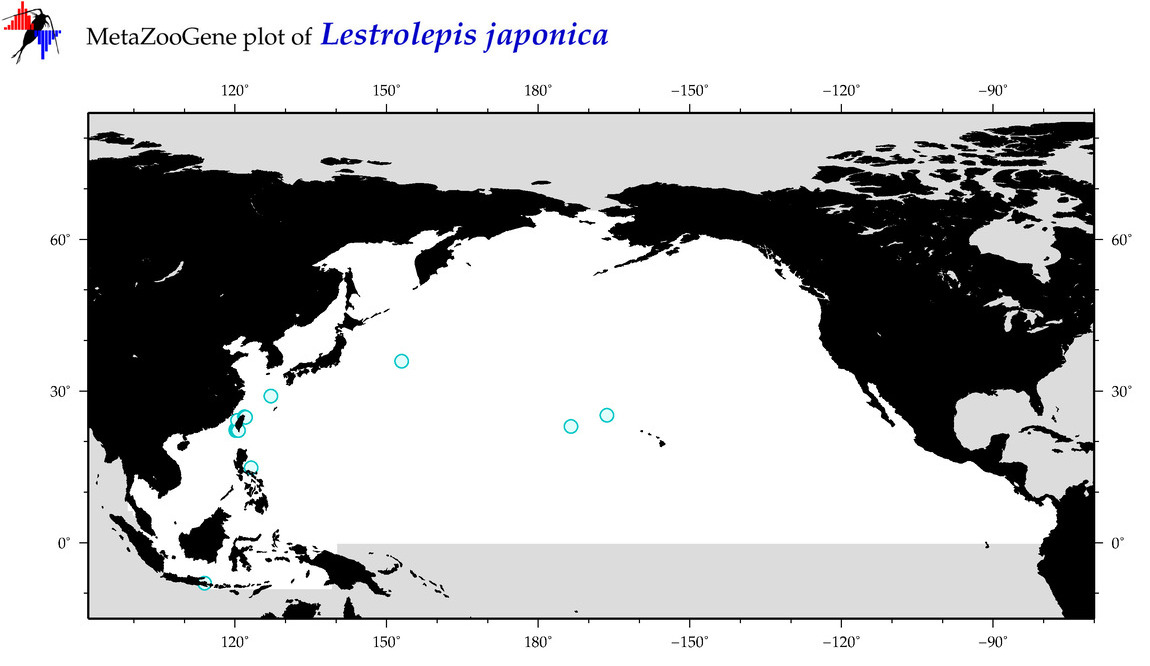 |
accepted
T5001998
R:1:0:0:0 |
| Lestrolepis luetkeni |
Species
(59) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028220
R:1:0:0:0 |
| Lestrolepis pofi |
Species
(60) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021574
R:1:0:0:0 |
| Macroparalepis johnfitchi |
Species
(61) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 2
18S = 0
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004607
R:1:0:0:0 |
| Magnisudis atlantica |
Species
(62) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
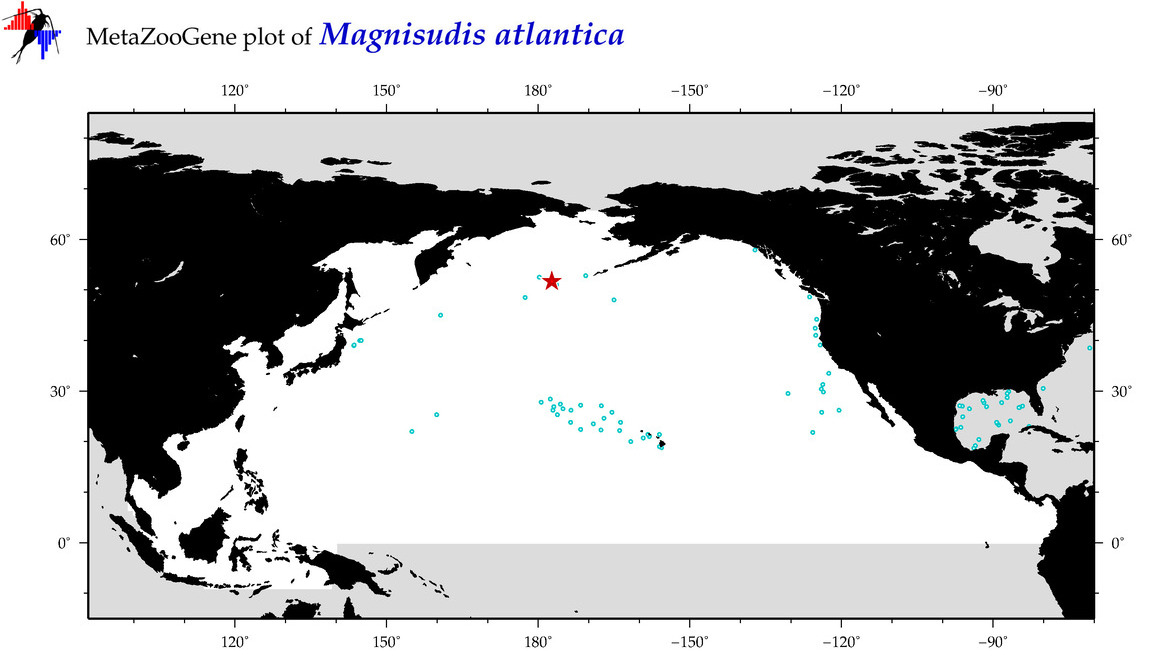 |
accepted
T5000861
R:1:0:0:0 |
| Magnisudis indica |
Species
(63) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021952
R:1:0:0:0 |
| Odontostomops normalops |
Species
(64) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
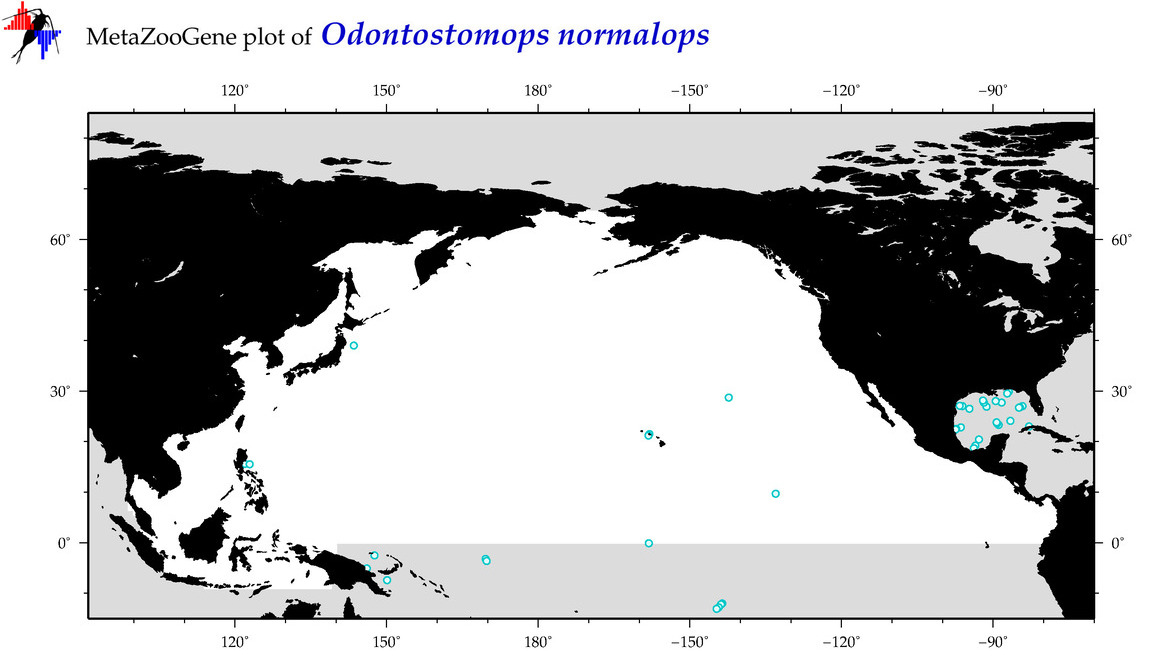 |
accepted
T5001002
R:1:0:0:0 |
| Omosudis lowii |
Species
(65) |
COI
12S
16S
18S
28S
|
COI = 27
12S = 5
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
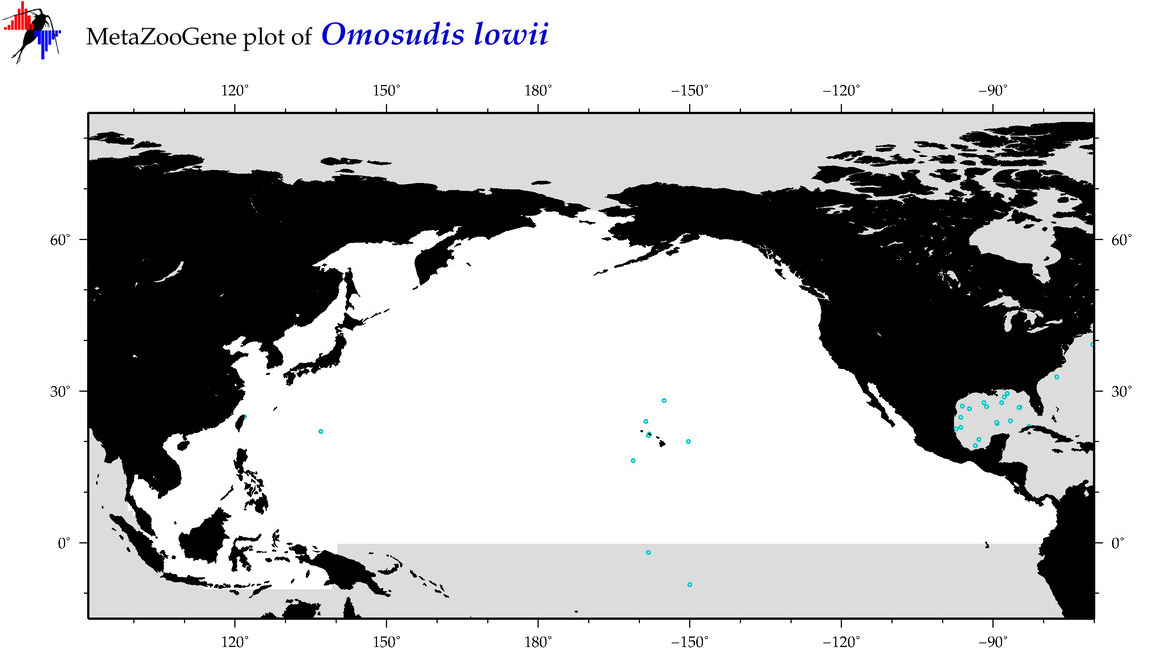 |
accepted
T5001009
R:1:0:0:0 |
| Paraulopus brevirostris |
Species
(66) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023673
R:1:0:0:0 |
| Paraulopus filamentosus |
Species
(67) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023674
R:1:0:0:0 |
| Paraulopus oblongus |
Species
(68) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
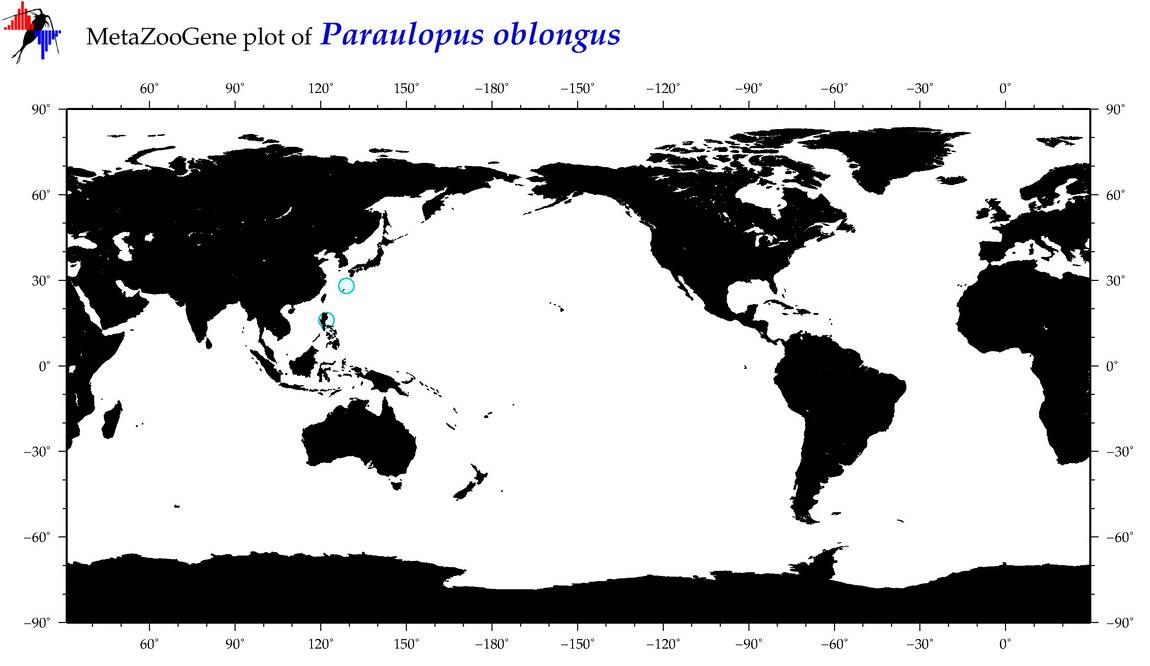 |
accepted
T5011186
R:1:0:0:0 |
| Rosenblattichthys alatus |
Species
(69) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 2
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025021
R:1:0:0:0 |
| Rosenblattichthys hubbsi |
Species
(70) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001988
R:1:0:0:0 |
| Rosenblattichthys volucris |
Species
(71) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001989
R:1:0:0:0 |
| Saurida argentea |
Species
(72) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005346
R:1:0:0:0 |
| Saurida brasiliensis |
Species
(73) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
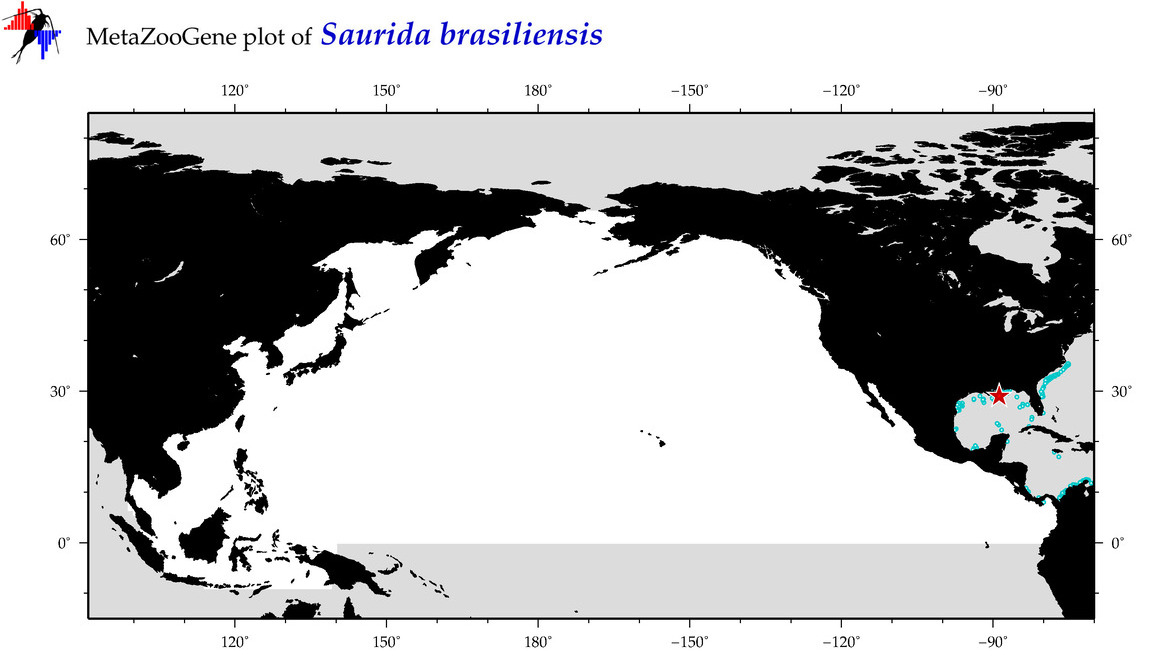 |
accepted
T5001192
R:1:0:0:0 |
| Saurida elongata |
Species
(74) |
COI
12S
16S
18S
28S
|
COI = 48
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 36
12S = 0
16S = 0
18S = 0
28S = 0
|
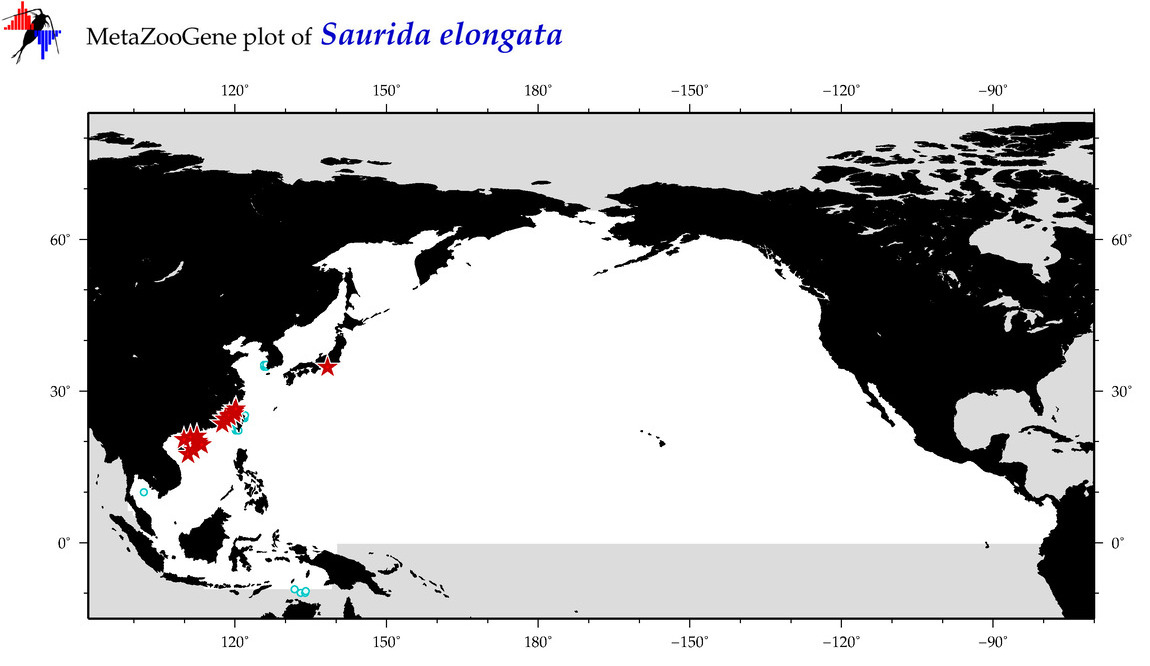 |
accepted
T5005348
R:1:0:0:0 |
| Saurida filamentosa |
Species
(75) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
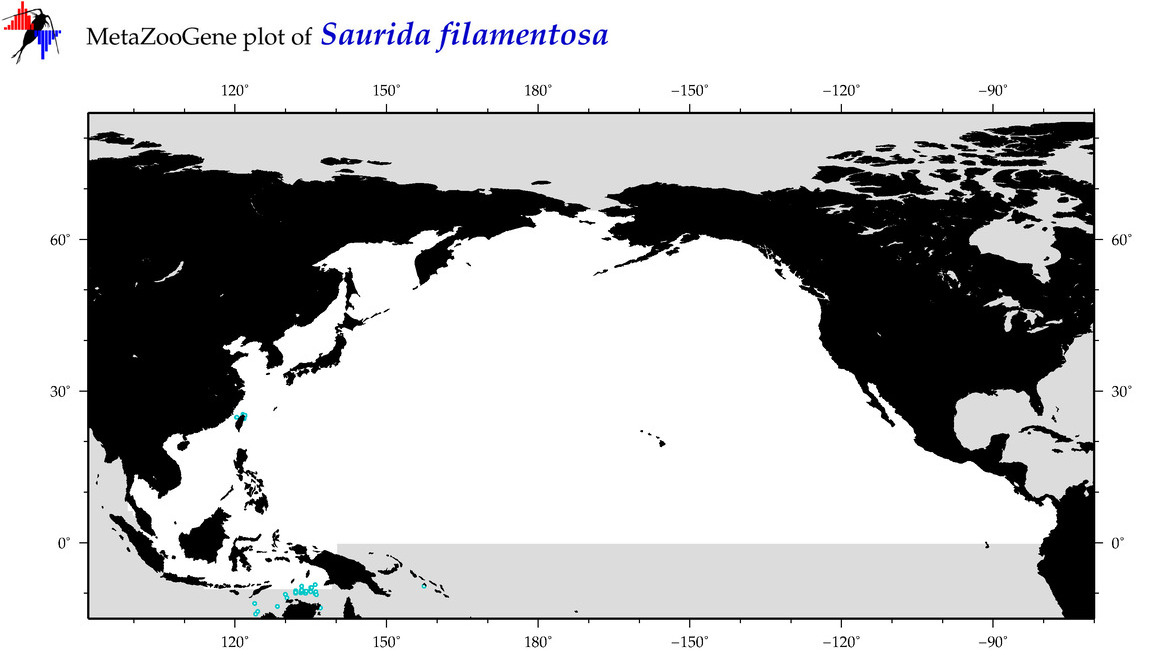 |
accepted
T5005349
R:1:0:0:0 |
| Saurida flamma |
Species
(76) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
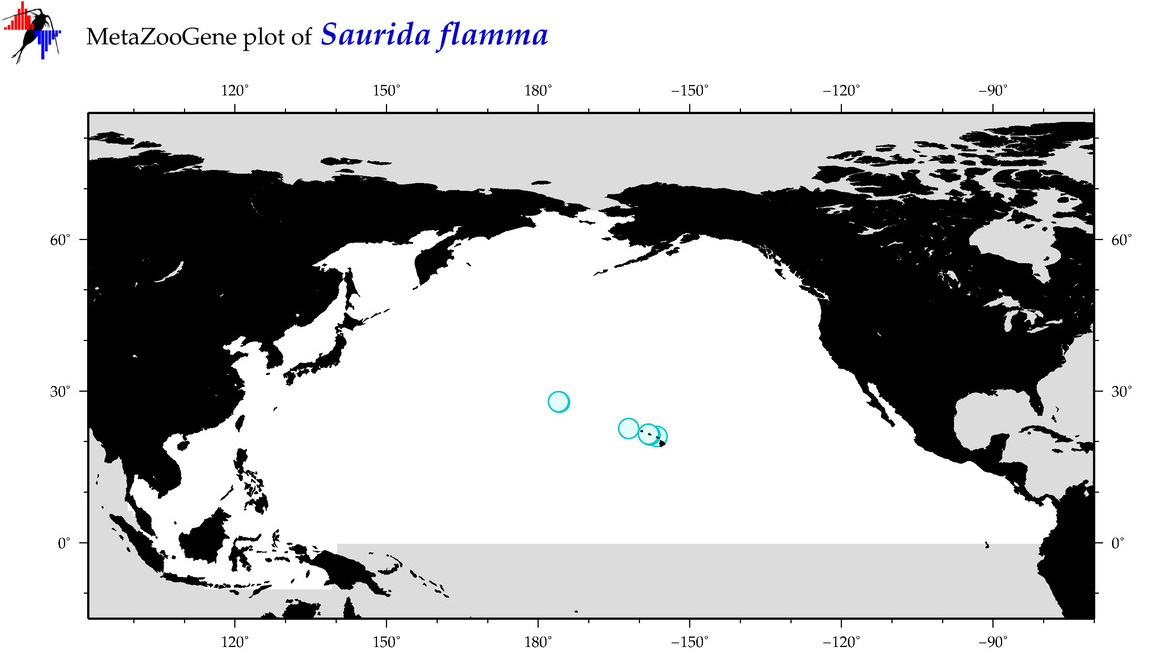 |
accepted
T5005350
R:1:0:0:0 |
| Saurida gracilis |
Species
(77) |
COI
12S
16S
18S
28S
|
COI = 27
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
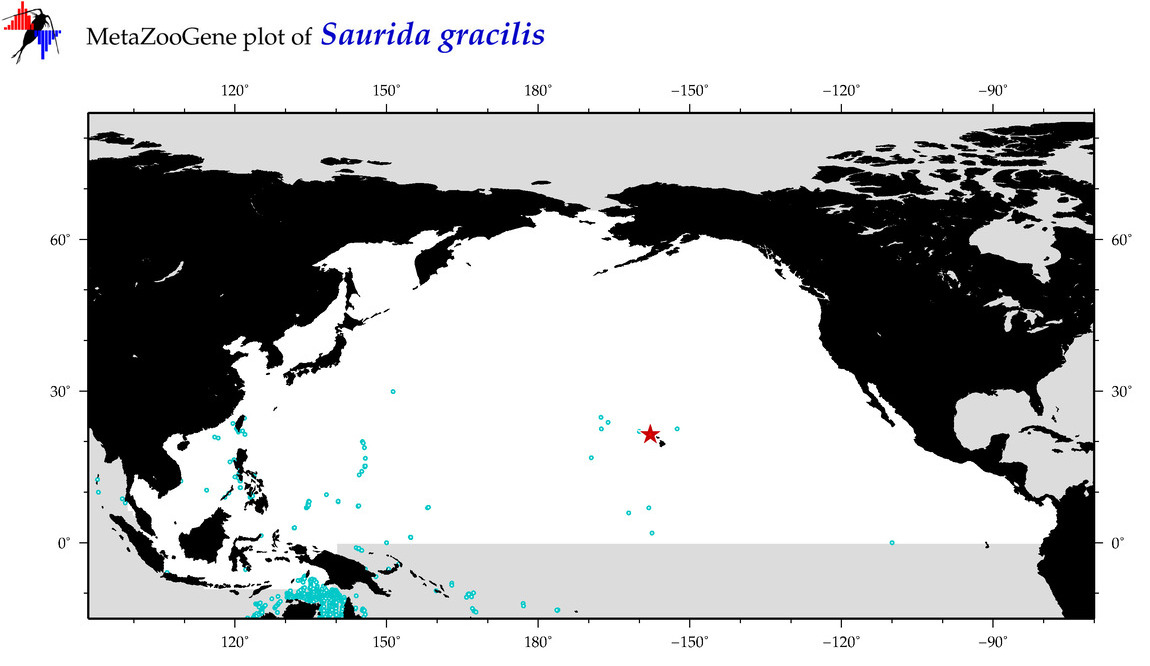 |
accepted
T5005351
R:1:0:0:0 |
| Saurida isarankurai |
Species
(78) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
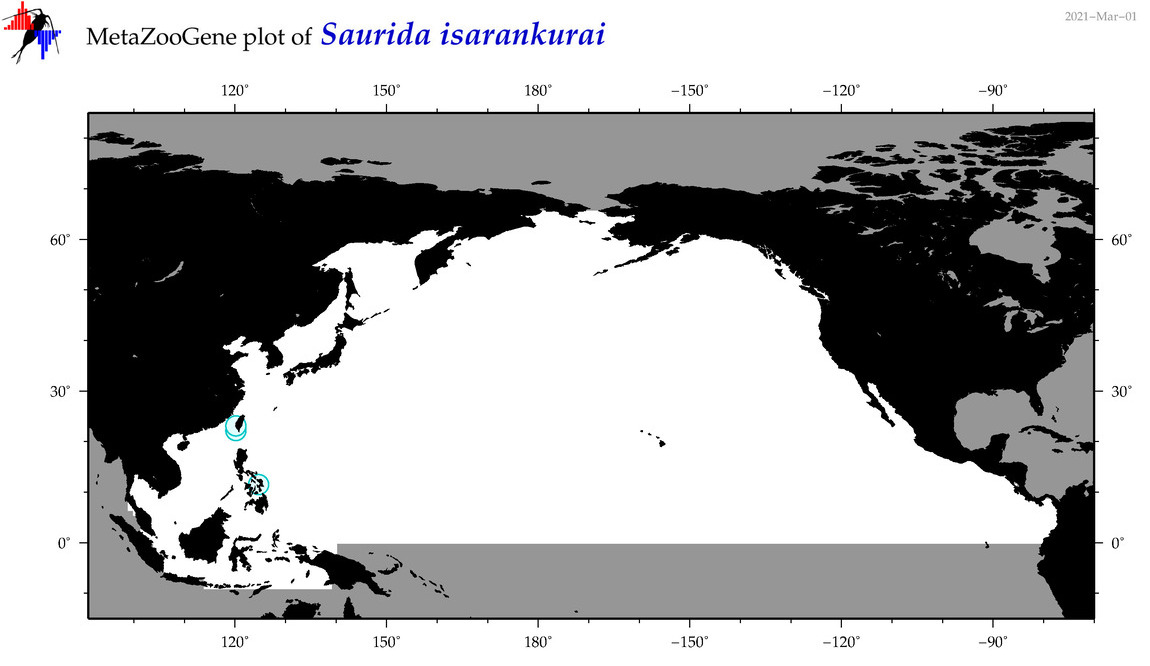 |
accepted
T5005353
R:1:0:0:0 |
| Saurida longimanus |
Species
(79) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 0
16S = 8
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005355
R:1:0:0:0 |
| Saurida macrolepis |
Species
(80) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 5
16S = 1
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005356
R:1:0:0:0 |
| Saurida micropectoralis |
Species
(81) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
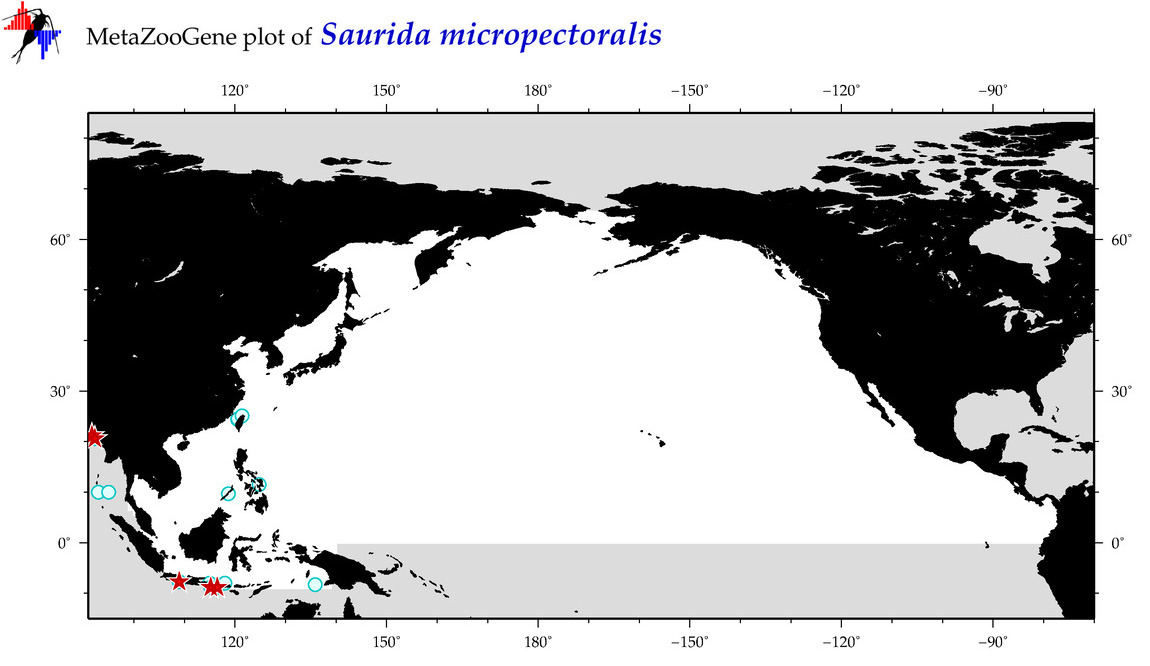 |
accepted
T5005358
R:1:0:0:0 |
| Saurida nebulosa |
Species
(82) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 3
16S = 3
18S = 0
28S = 0
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
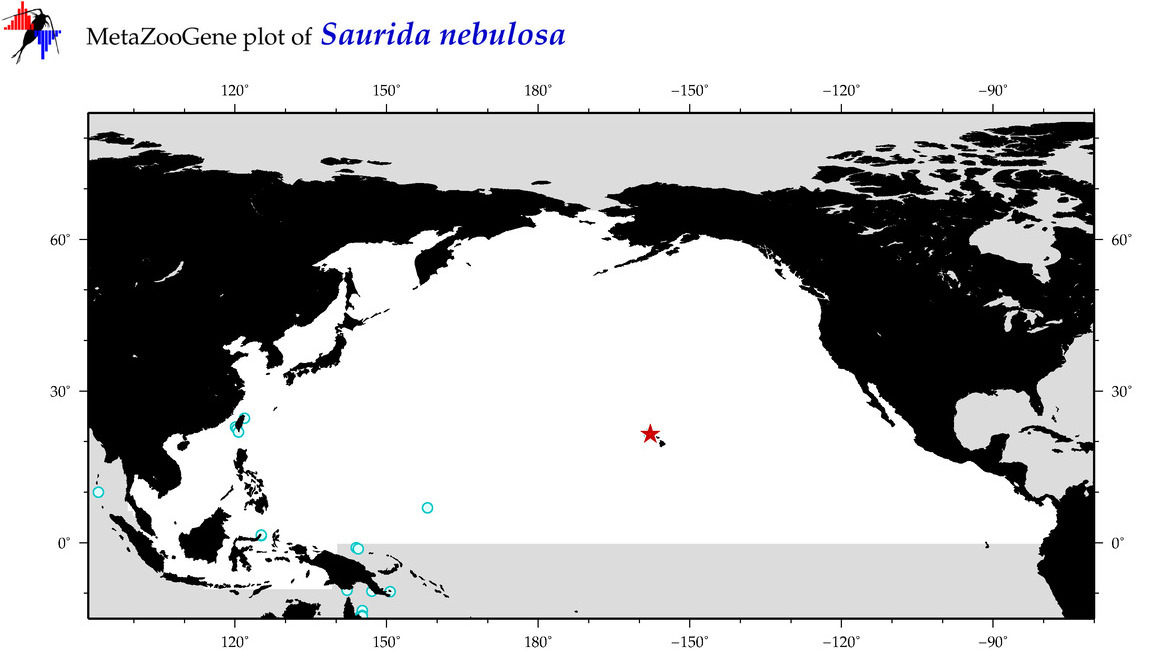 |
accepted
T5005359
R:1:1:0:0 |
| Saurida tumbil |
Species
(83) |
COI
12S
16S
18S
28S
|
COI = 53
12S = 1
16S = 6
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
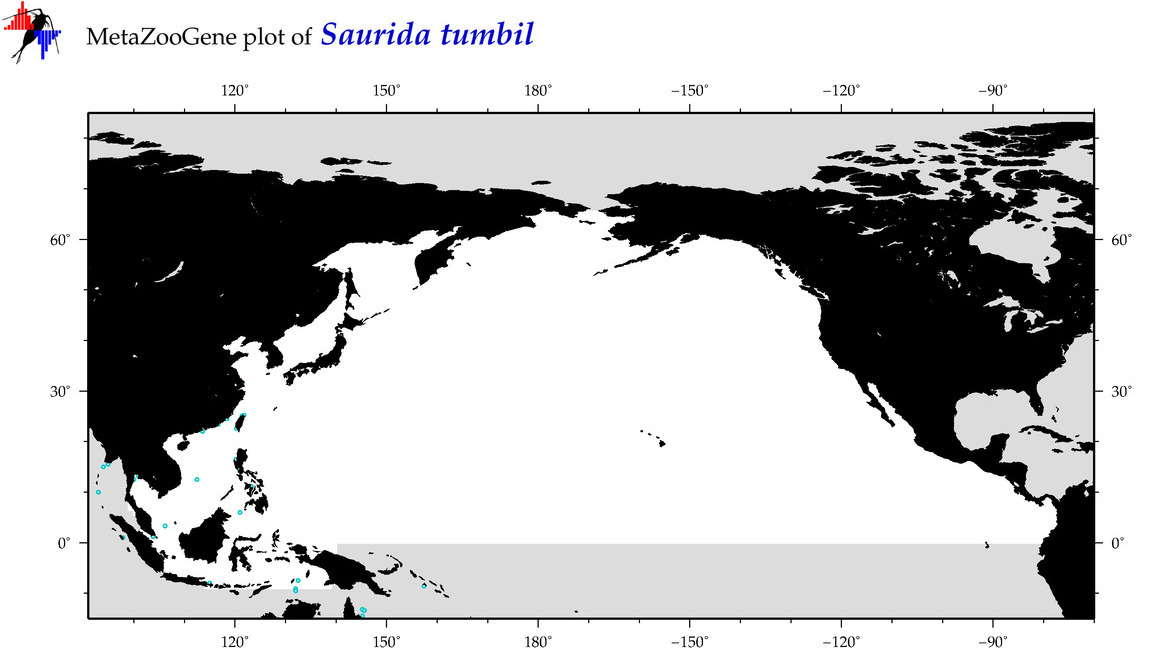 |
accepted
T5005362
R:1:0:0:0 |
| Saurida umeyoshii |
Species
(84) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 4
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
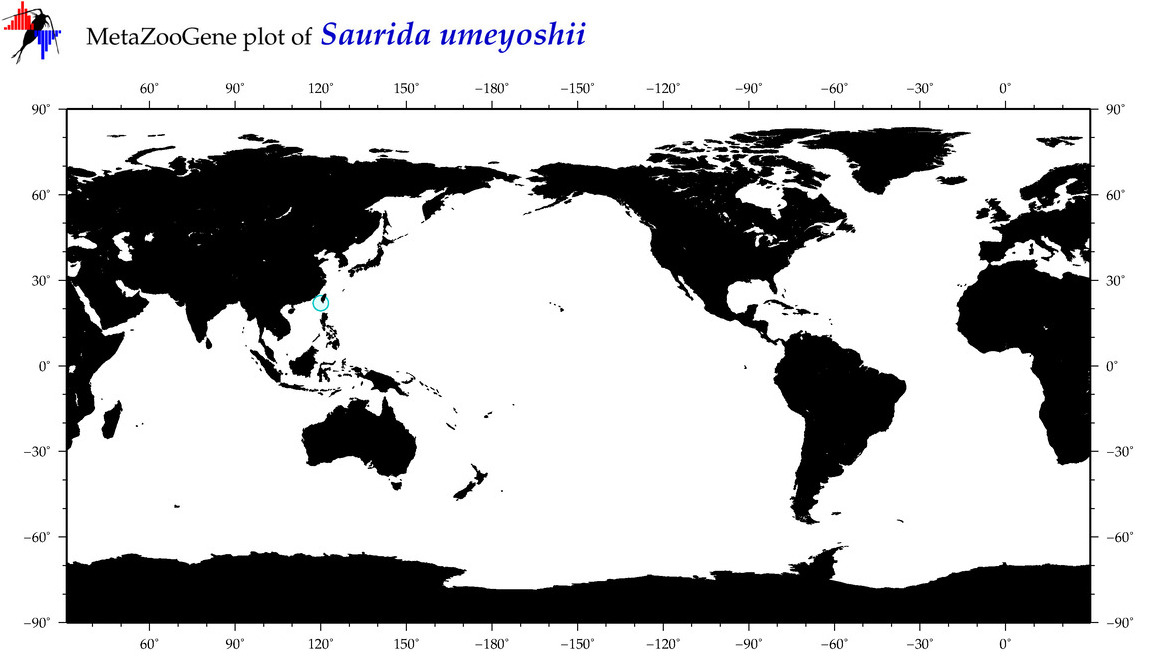 |
accepted
T5005363
R:1:0:0:0 |
| Saurida undosquamis |
Species
(85) |
COI
12S
16S
18S
28S
|
COI = 129
12S = 4
16S = 10
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005364
R:1:0:0:0 |
| Saurida wanieso |
Species
(86) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 9
16S = 7
18S = 0
28S = 0
|
COI = 4
12S = 2
16S = 2
18S = 0
28S = 0
|
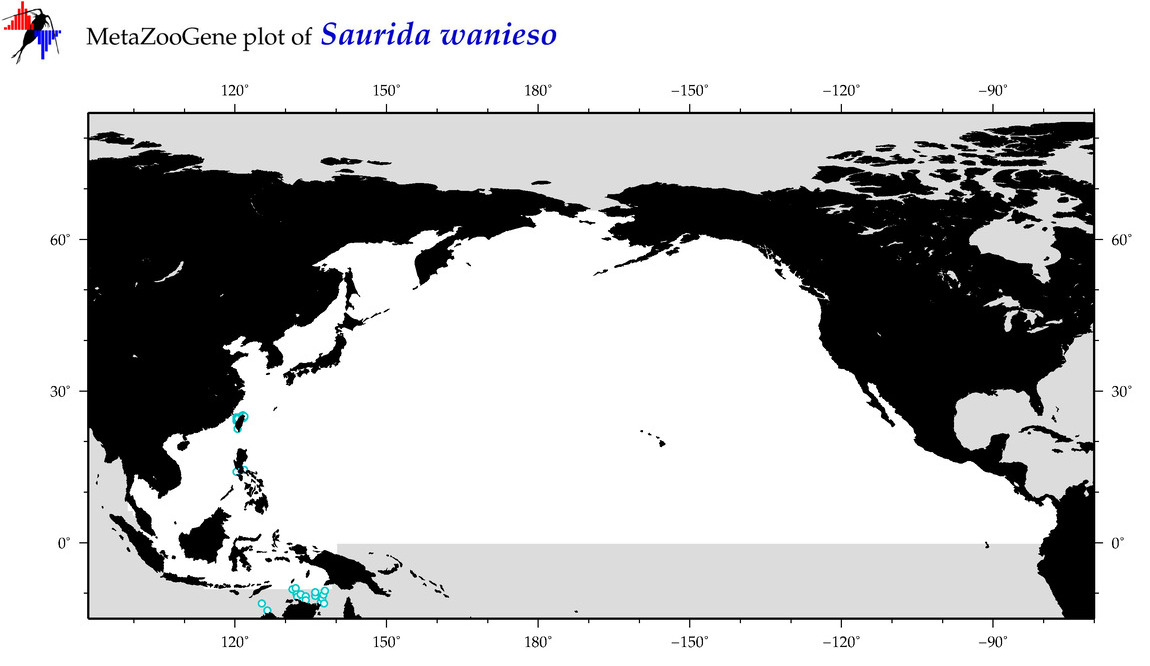 |
accepted
T5005365
R:1:0:0:0 |
| Scopelarchoides danae |
Species
(87) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005376
R:1:0:0:0 |
| Scopelarchoides nicholsi |
Species
(88) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001212
R:1:0:0:0 |
| Scopelarchoides signifer |
Species
(89) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025317
R:1:0:0:0 |
| Scopelarchus analis |
Species
(90) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
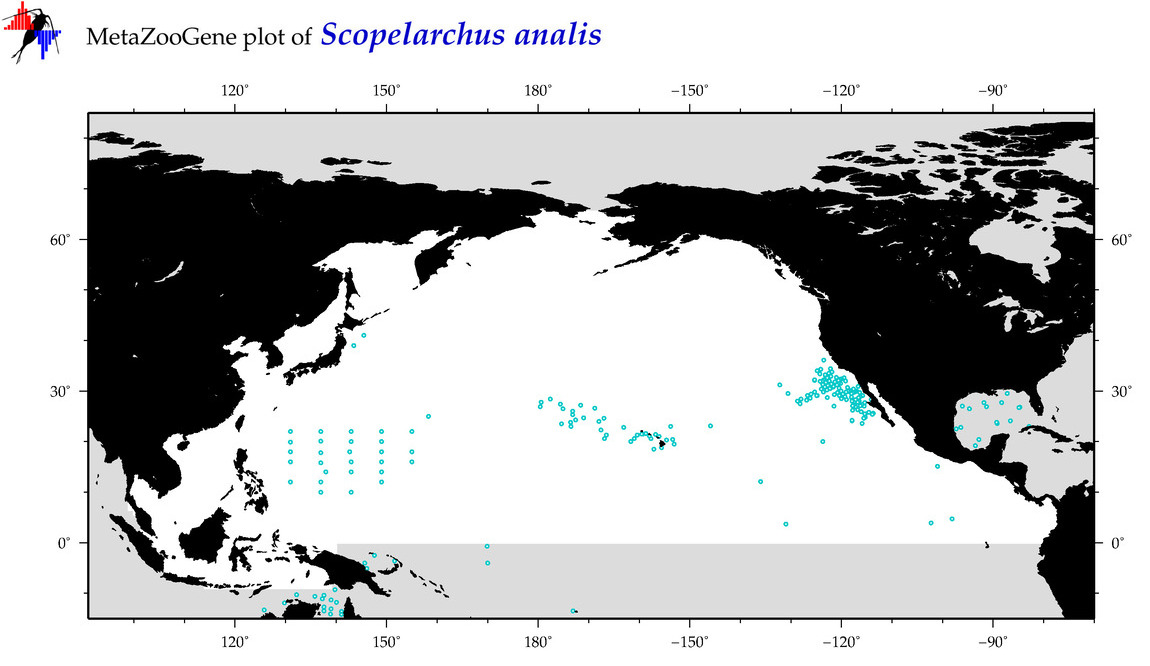 |
accepted
T5001214
R:1:0:0:0 |
| Scopelarchus guentheri |
Species
(91) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
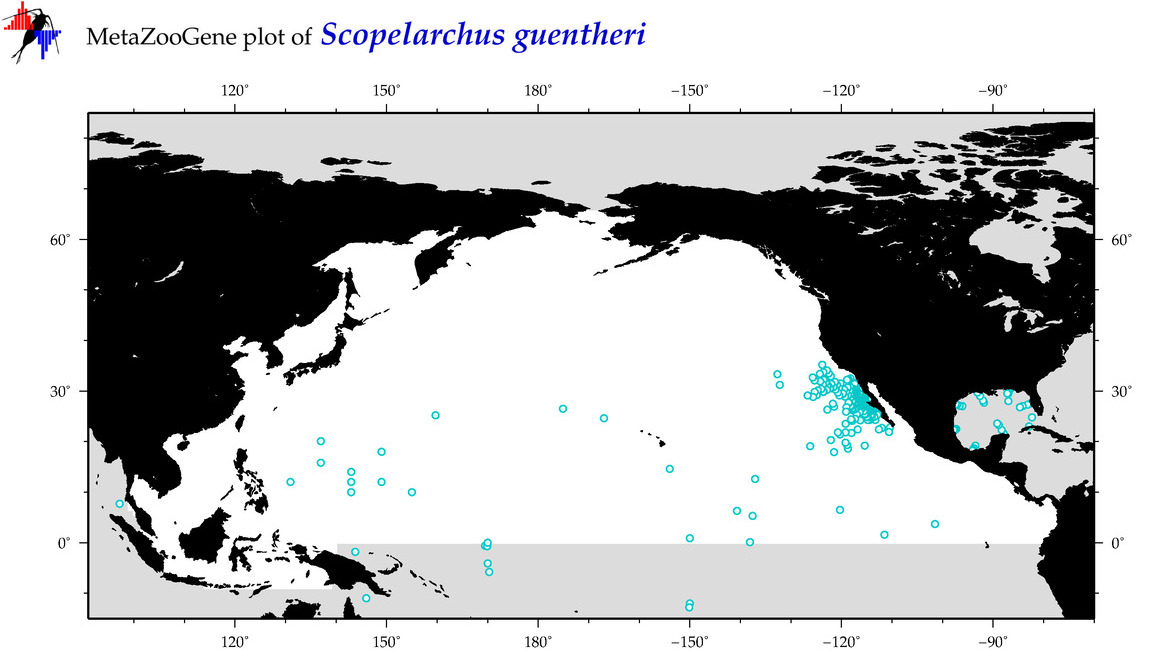 |
accepted
T5001215
R:1:0:0:0 |
| Scopelarchus michaelsarsi |
Species
(92) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
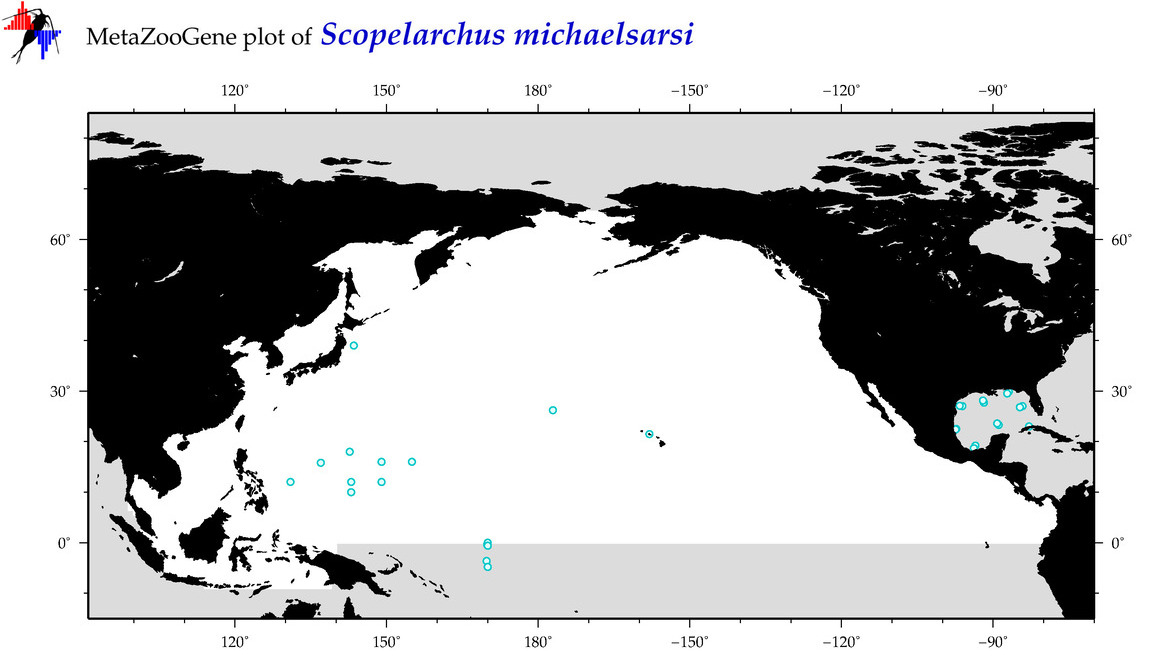 |
accepted
T5001216
R:1:0:0:0 |
| Scopelarchus stephensi |
Species
(93) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
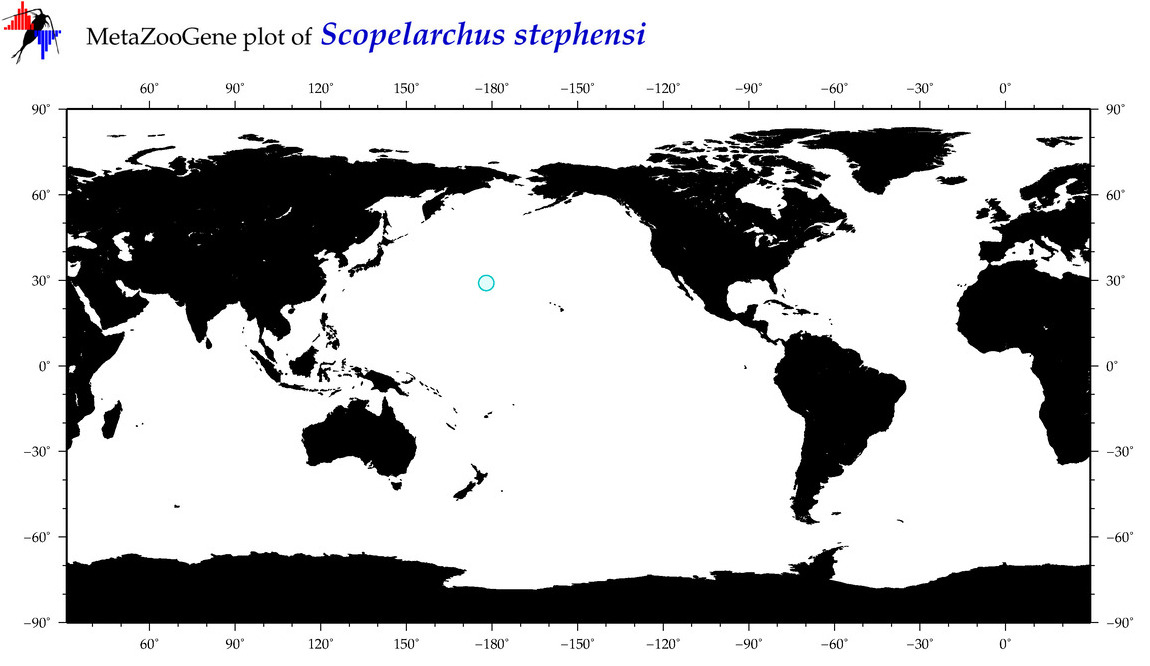 |
accepted
T5005378
R:1:0:0:0 |
| Scopelosaurus adleri |
Species
(94) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5025323
R:1:0:0:0 |
| Scopelosaurus ahlstromi |
Species
(95) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005383
R:1:0:0:0 |
| Scopelosaurus harryi |
Species
(96) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
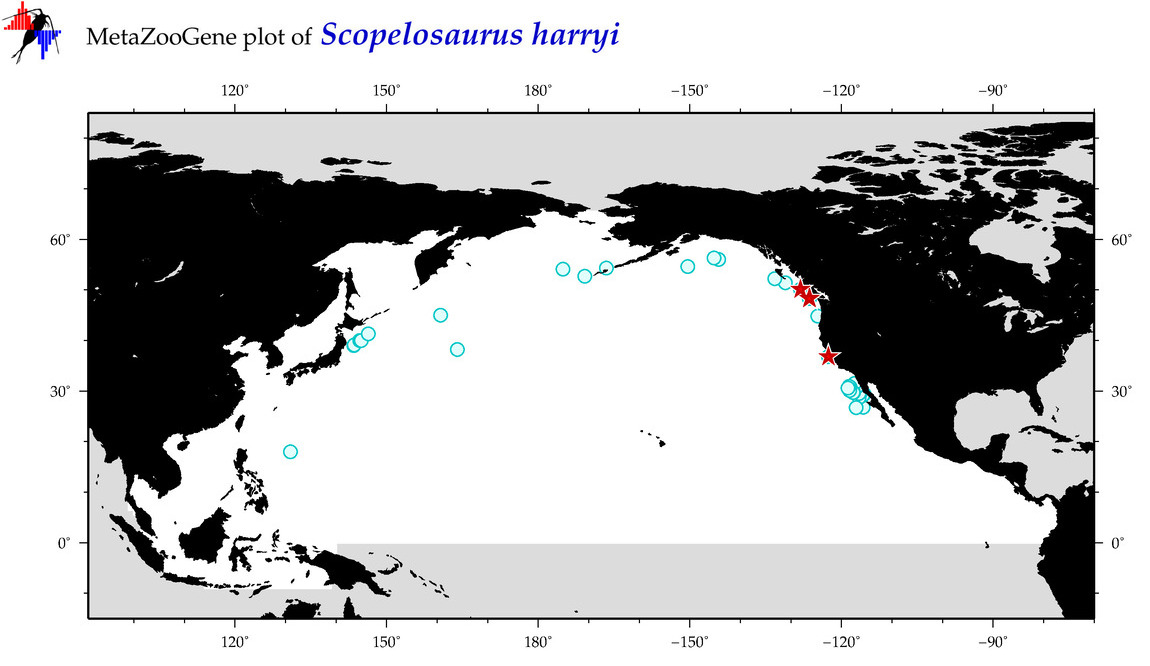 |
accepted
T5001223
R:1:0:0:0 |
| Scopelosaurus hoedti |
Species
(97) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001829
R:1:0:0:0 |
| Scopelosaurus hubbsi |
Species
(98) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001999
R:1:0:0:0 |
| Scopelosaurus mauli |
Species
(99) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001224
R:1:0:0:0 |
| Scopelosaurus smithii |
Species
(100) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
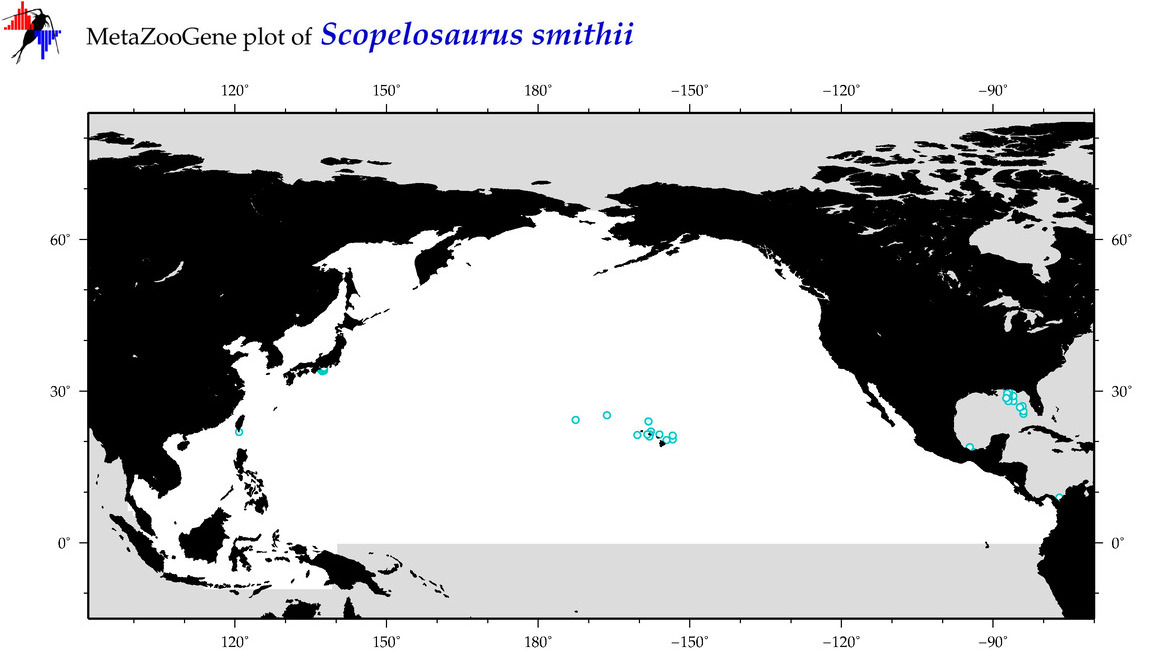 |
accepted
T5001225
R:1:0:0:0 |
| Stemonosudis elegans |
Species
(101) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5002000
R:1:0:0:0 |
| Stemonosudis macrura |
Species
(102) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001993
R:1:0:0:0 |
| Stemonosudis rothschildi |
Species
(103) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
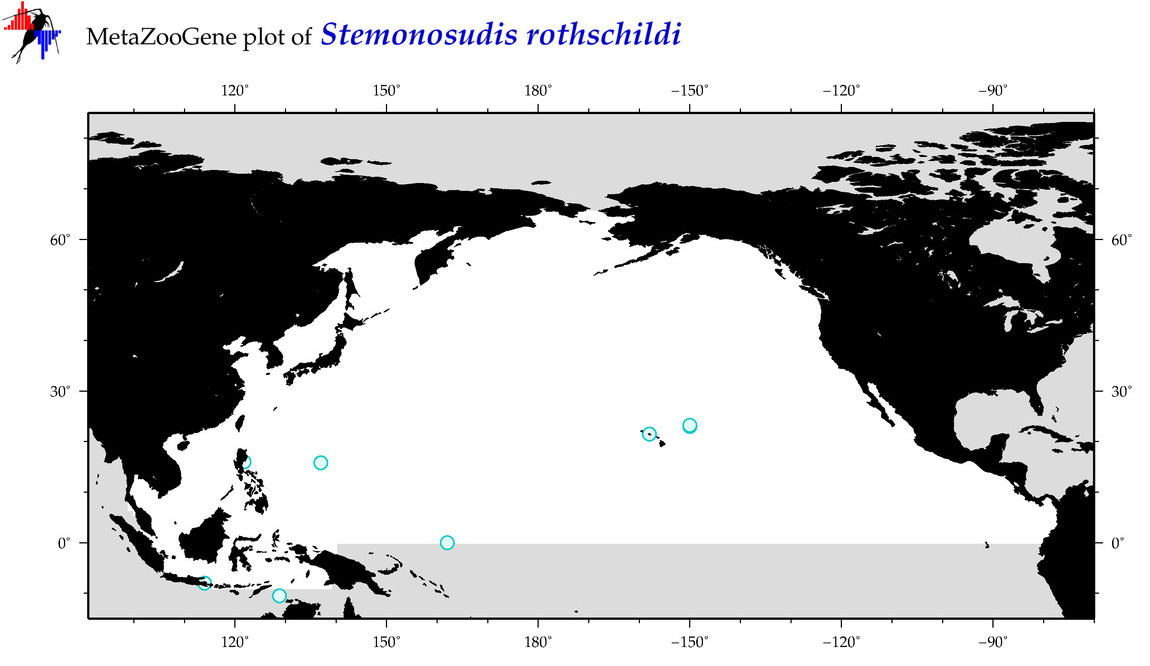 |
accepted
T5005607
R:1:0:0:0 |
| Sudis atrox |
Species
(104) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
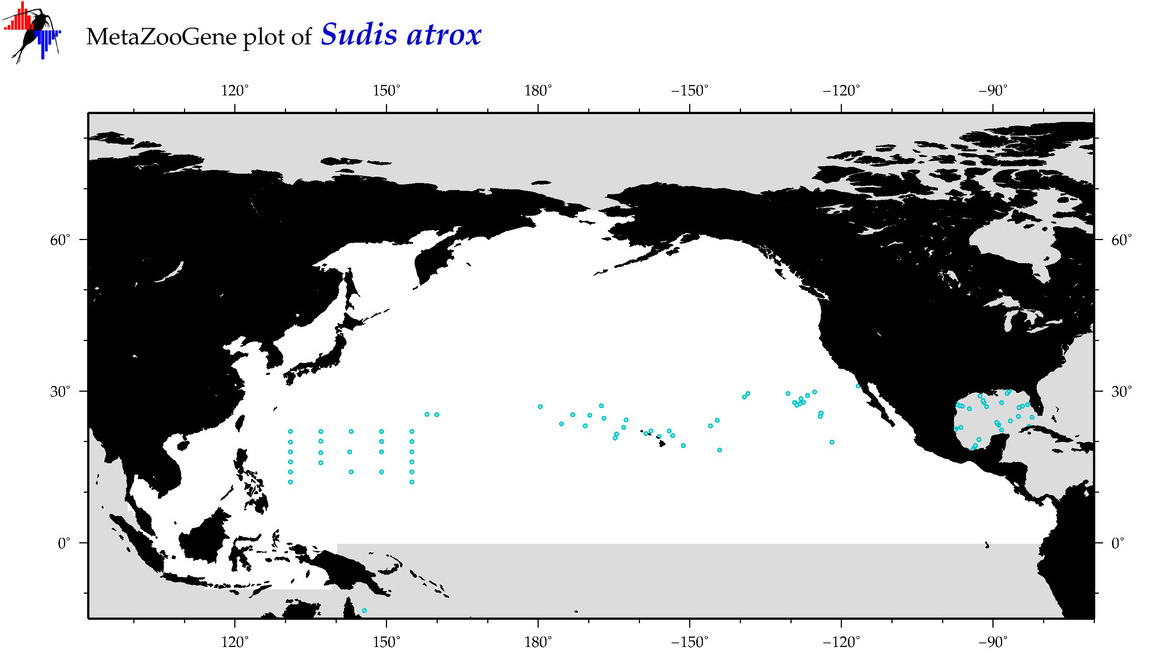 |
accepted
T5001304
R:1:0:0:0 |
| Sudis hyalina |
Species
(105) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001305
R:1:0:0:0 |
| Synodus binotatus |
Species
(106) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001825
R:1:0:0:0 |
| Synodus capricornis |
Species
(107) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005685
R:1:0:0:0 |
| Synodus dermatogenys |
Species
(108) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
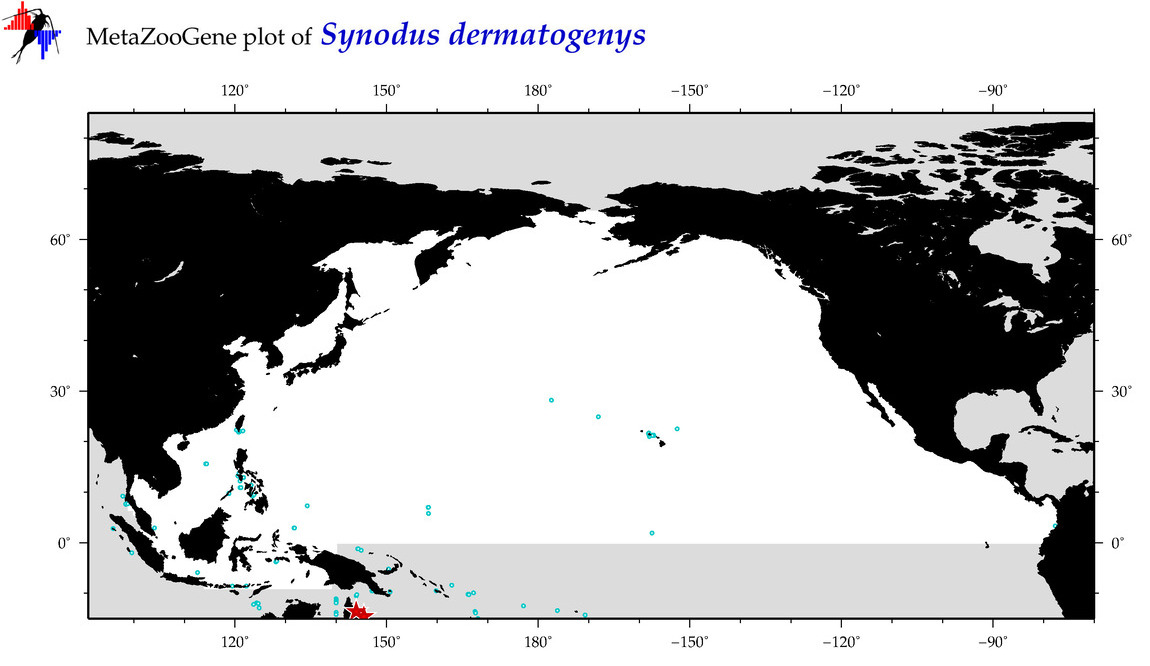 |
accepted
T5005686
R:1:0:0:0 |
| Synodus doaki |
Species
(109) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005687
R:1:0:0:0 |
| Synodus evermanni |
Species
(110) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001994
R:1:0:0:0 |
| Synodus falcatus |
Species
(111) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026086
R:1:0:0:0 |
| Synodus fuscus |
Species
(112) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026087
R:1:0:0:0 |
| Synodus hoshinonis |
Species
(113) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 5
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
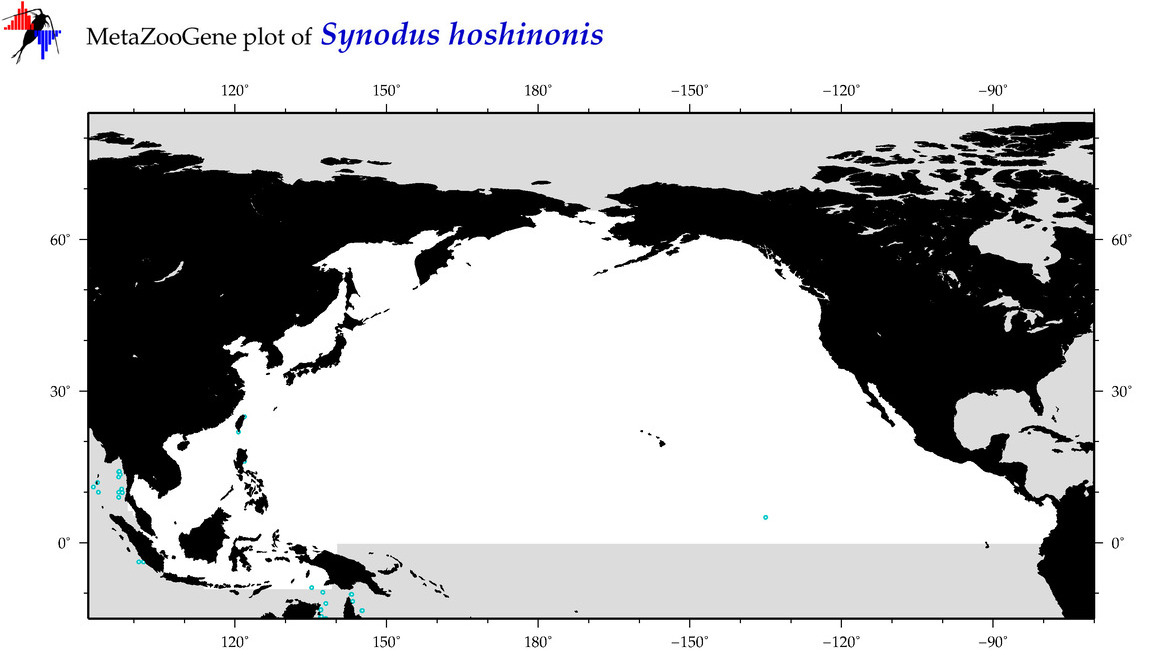 |
accepted
T5005688
R:1:1:0:0 |
| Synodus indicus |
Species
(114) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005689
R:1:0:0:0 |
| Synodus jaculum |
Species
(115) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005691
R:1:0:0:0 |
| Synodus kaianus |
Species
(116) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005692
R:1:0:0:0 |
| Synodus lacertinus |
Species
(117) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005693
R:1:0:0:0 |
| Synodus lobeli |
Species
(118) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026089
R:1:0:0:0 |
| Synodus lucioceps |
Species
(119) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
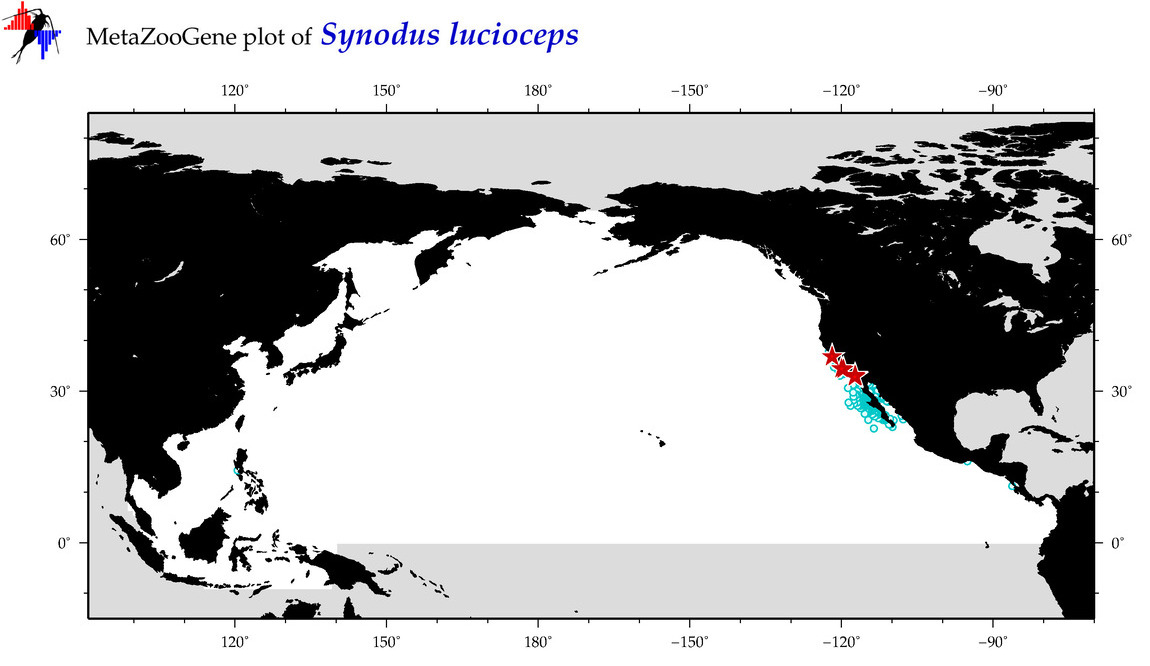 |
accepted
T5001329
R:1:0:0:0 |
| Synodus macrops |
Species
(120) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
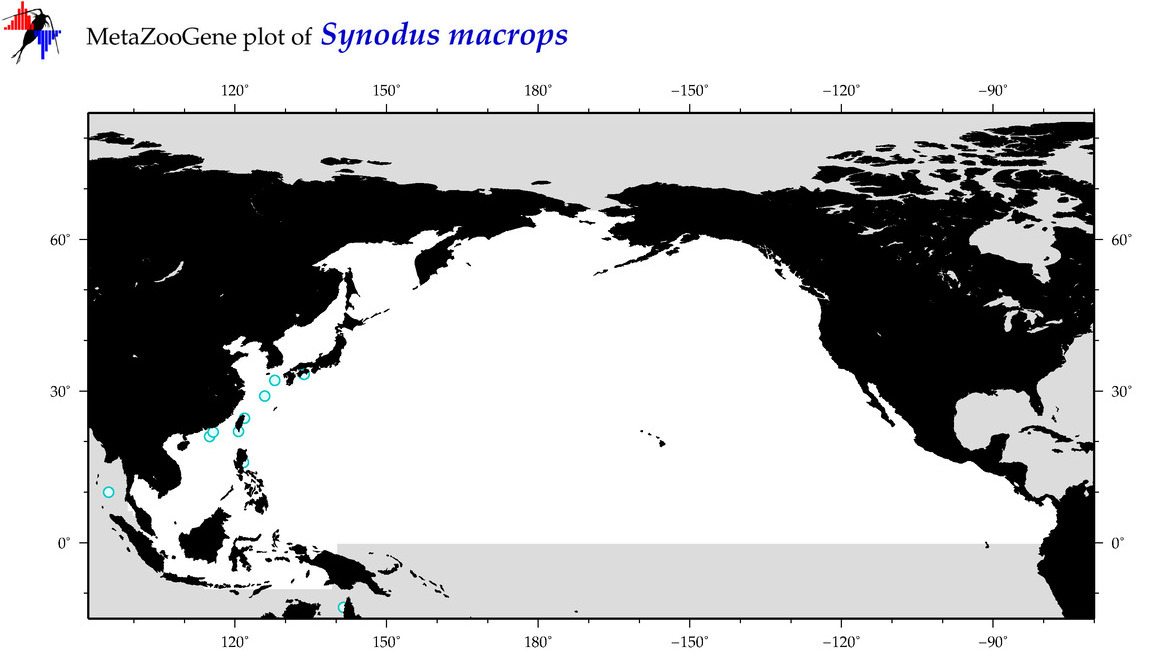 |
accepted
T5005694
R:1:0:0:0 |
| Synodus oculeus |
Species
(121) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026091
R:1:0:0:0 |
| Synodus rubromarmoratus |
Species
(122) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005697
R:1:0:0:0 |
| Synodus sageneus |
Species
(123) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
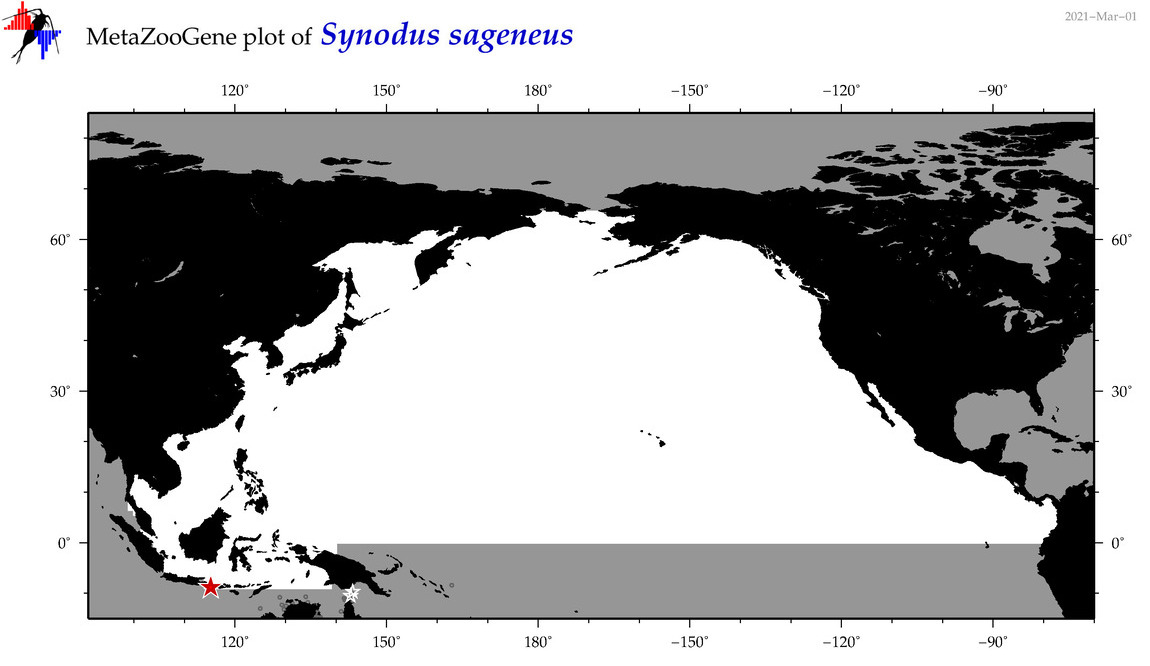 |
accepted
T5026093
R:1:0:0:0 |
| Synodus scituliceps |
Species
(124) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026094
R:1:0:0:0 |
| Synodus sechurae |
Species
(125) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5001995
R:1:0:0:0 |
| Synodus synodus |
Species
(126) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
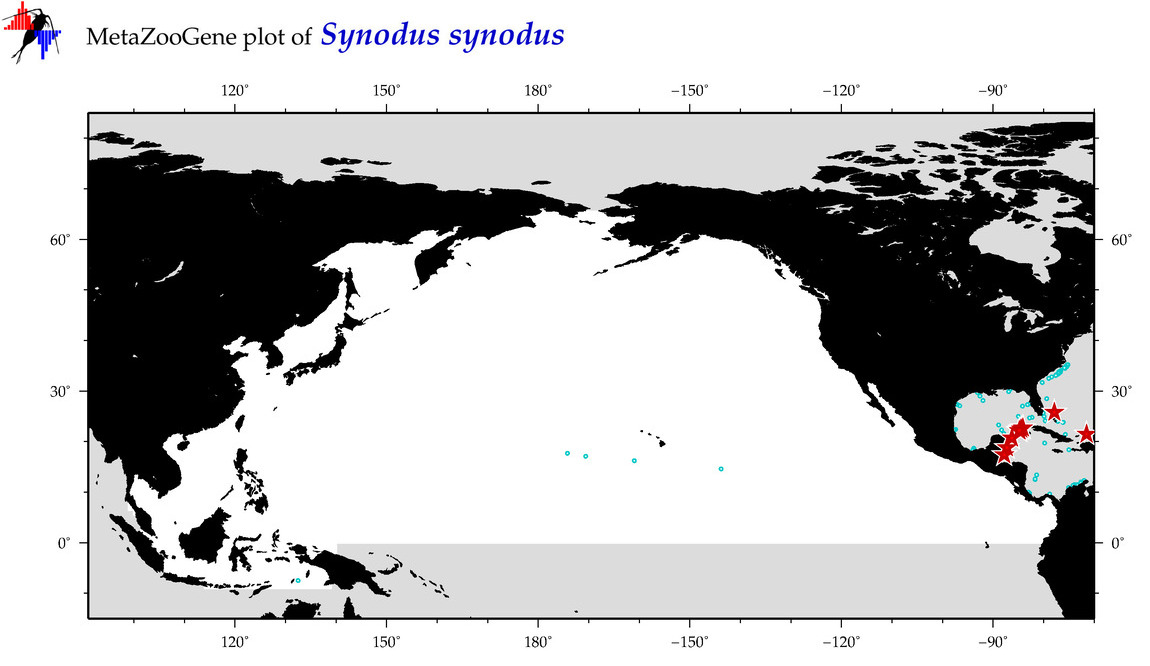 |
accepted
T5001330
R:1:0:0:0 |
| Synodus tectus |
Species
(127) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5026097
R:1:0:0:0 |
| Synodus ulae |
Species
(128) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 2
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5013256
R:1:0:0:0 |
| Synodus usitatus |
Species
(129) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005699
R:1:0:0:0 |
| Synodus variegatus |
Species
(130) |
COI
12S
16S
18S
28S
|
COI = 42
12S = 6
16S = 4
18S = 0
28S = 1
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
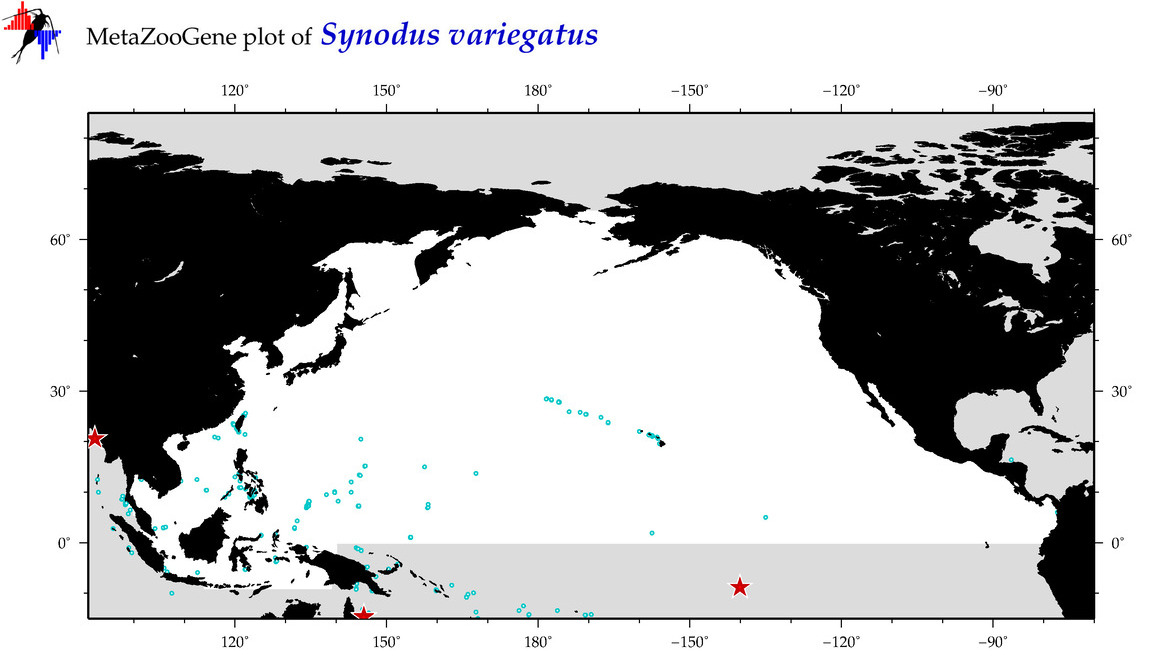 |
accepted
T5005700
R:1:0:0:0 |
| Trachinocephalus myops |
Species
(131) |
COI
12S
16S
18S
28S
|
COI = 111
12S = 10
16S = 3
18S = 0
28S = 0
|
COI = 32
12S = 0
16S = 0
18S = 0
28S = 0
|
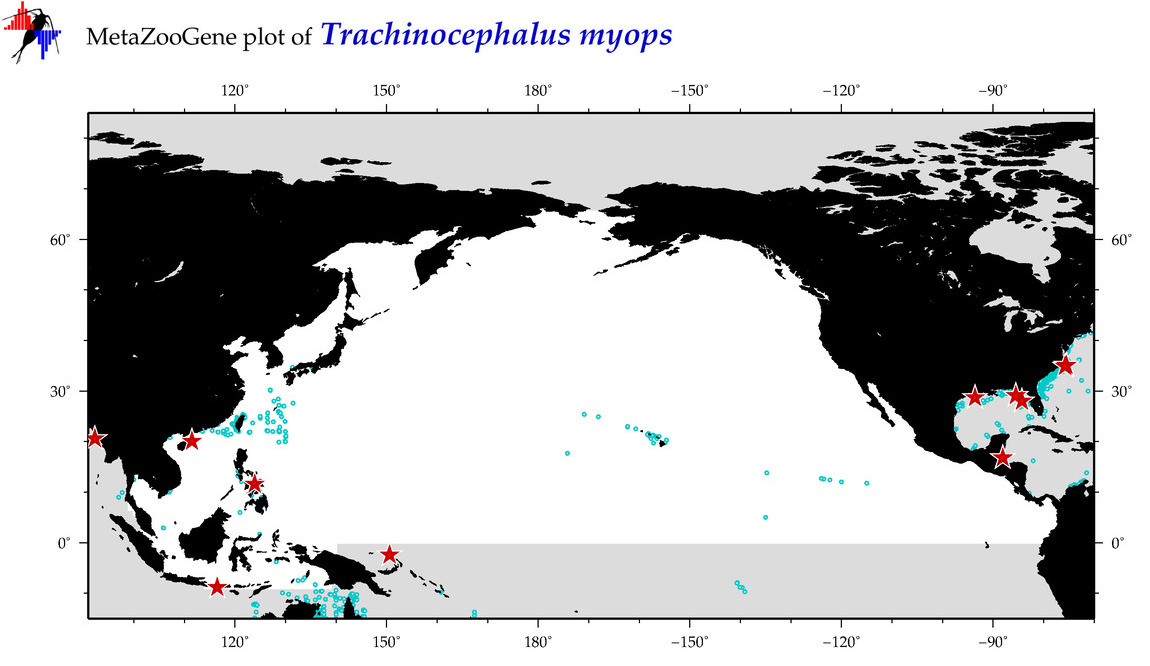 |
accepted
T5001354
R:1:0:0:0 |
| Uncisudis advena |
Species
(132) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
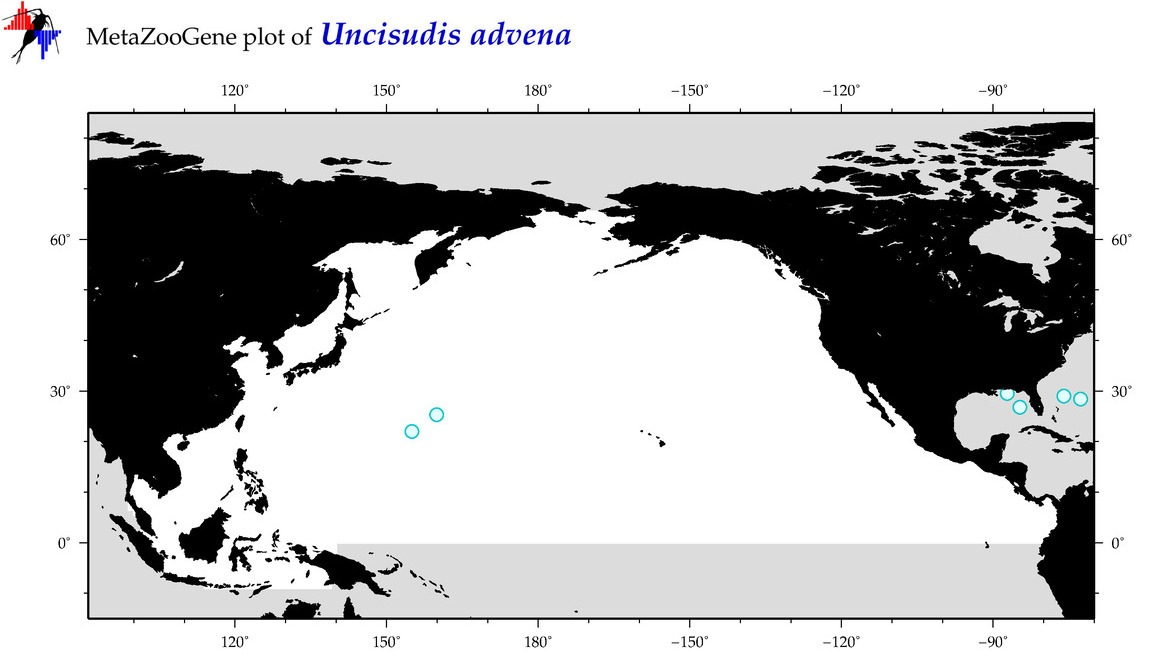 |
accepted
T5005813
R:1:0:0:0 |
| Uncisudis quadrimaculata |
Species
(133) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5013270
R:1:0:0:0 |