Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2023-m07-15
2023-Jul-21 |
| Abra alba |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 1
18S = 6
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
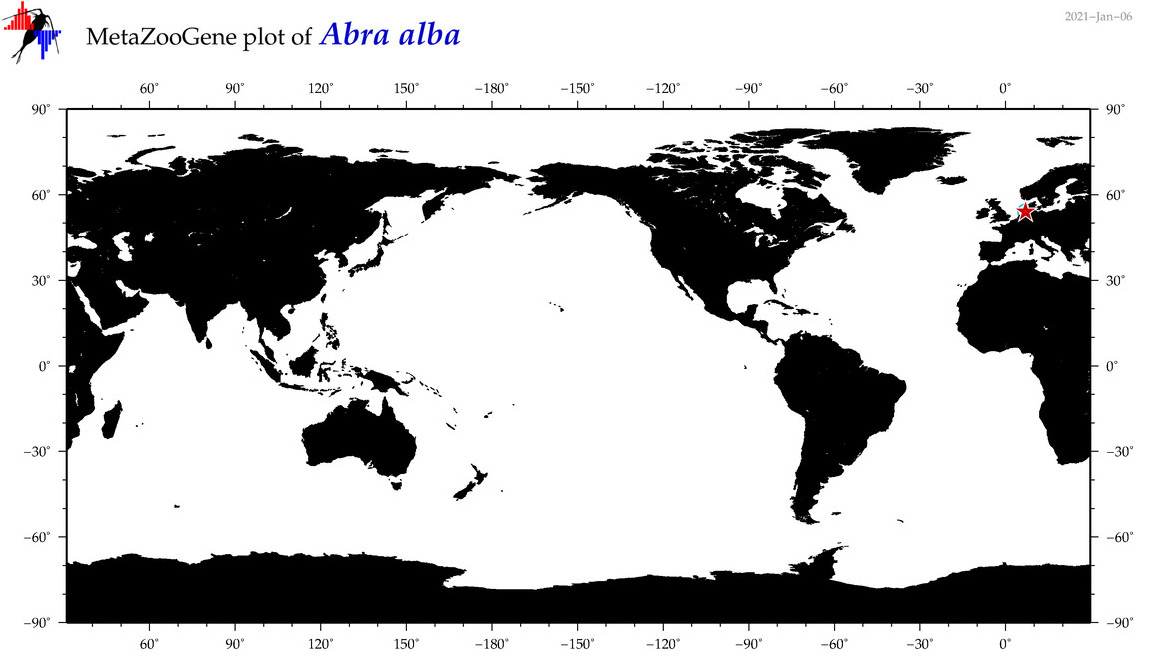 |
accepted
T4017135
R:1:0:0:0 |
| Abra nitida |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
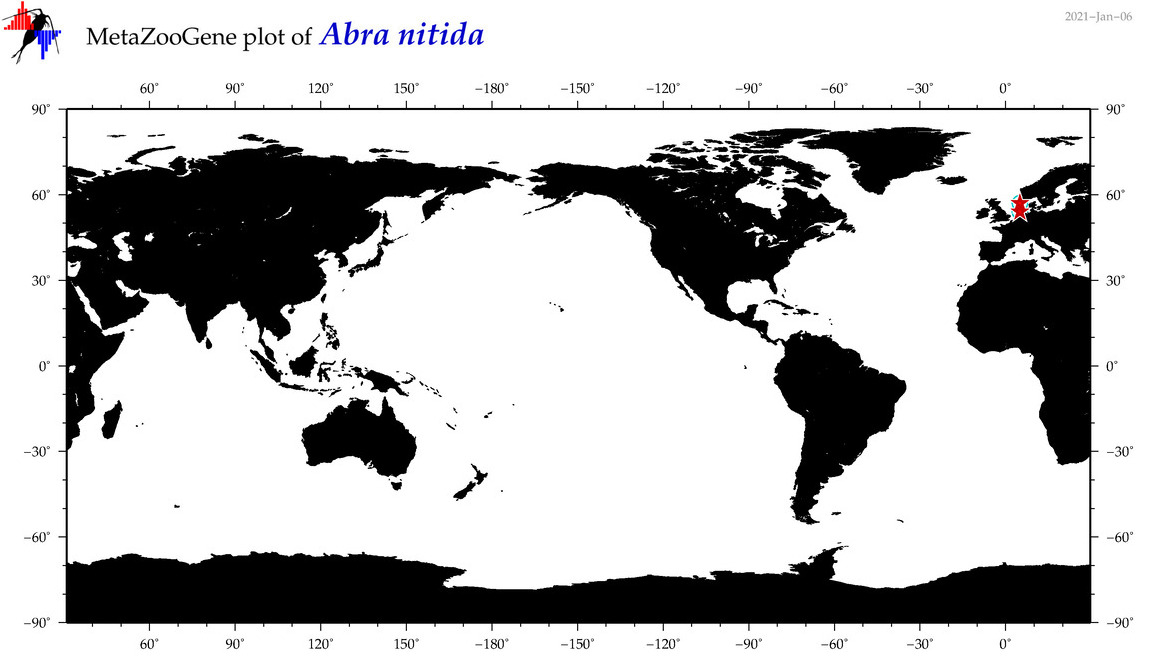 |
accepted
T4017136
R:1:0:0:0 |
| Abra prismatica |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
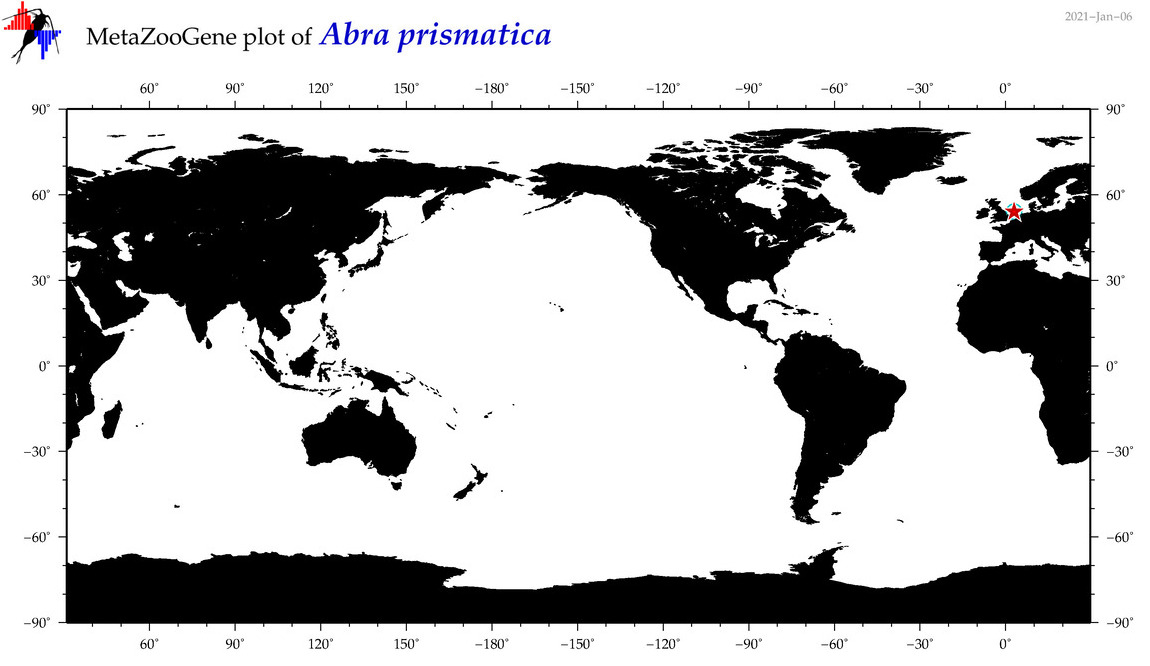 |
accepted
T4017137
R:1:0:0:0 |
| Acanthocardia echinata |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
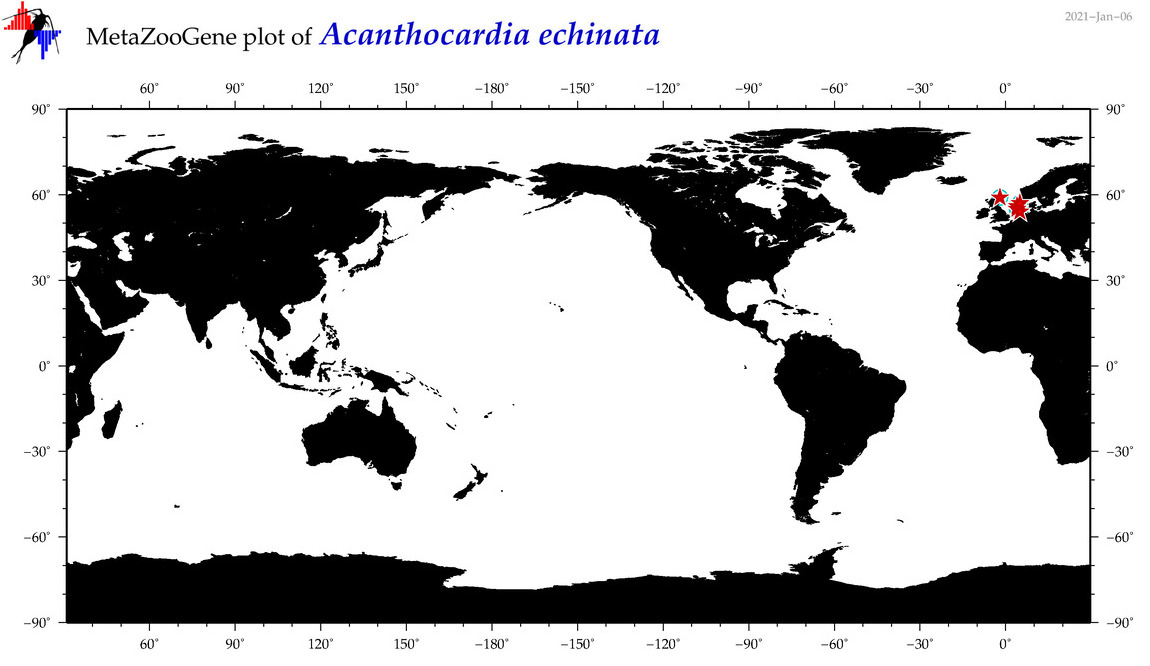 |
accepted
T4017074
R:1:0:0:0 |
| Aequipecten opercularis |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 4
16S = 13
18S = 8
28S = 5
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
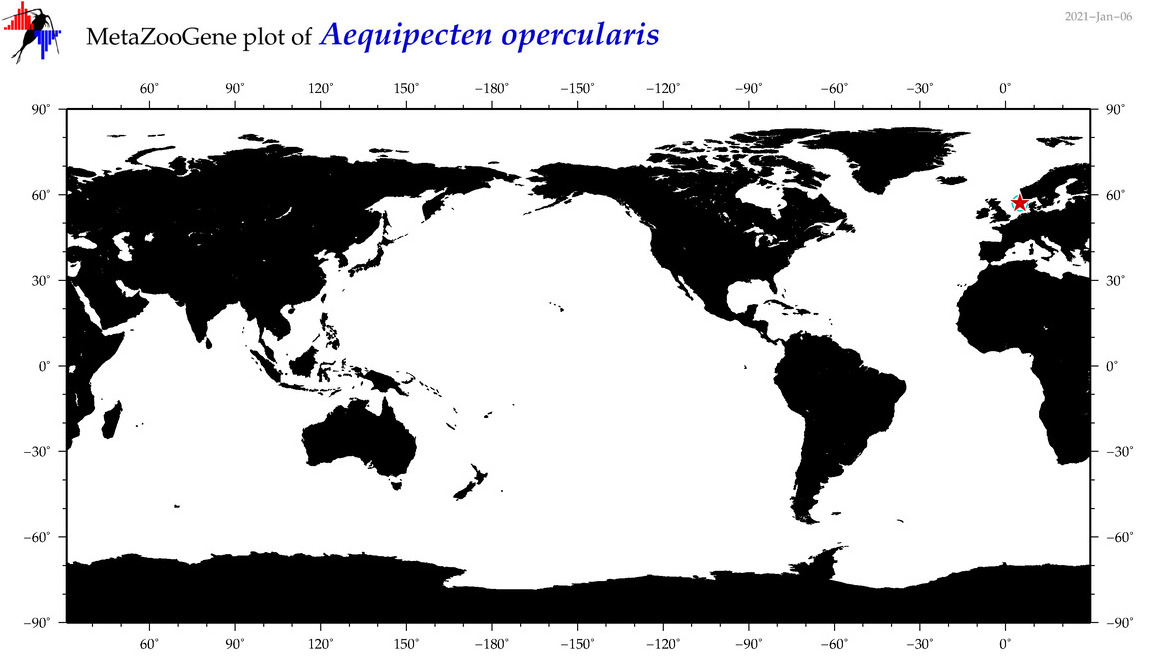 |
accepted
T4015897
R:1:0:0:0 |
| Anodonta anatina |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 584
12S = 12
16S = 125
18S = 0
28S = 89
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4133241
R:0:1:1:0 |
| Arctica islandica |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 22
12S = 3
16S = 3
18S = 4
28S = 4
|
COI = 2
12S = 1
16S = 1
18S = 0
28S = 0
|
 |
accepted
T4010115
R:1:0:0:0 |
| Asbjornsenia pygmaea |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
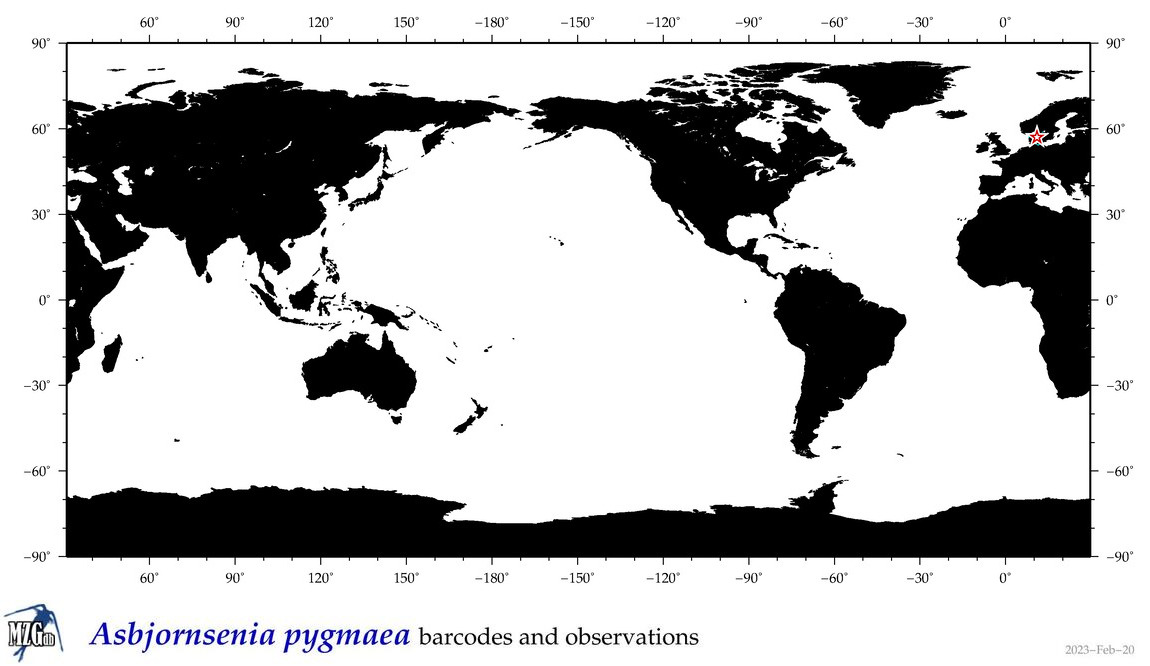 |
accepted
T4133313
R:1:0:0:0 |
| Astarte borealis |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 19
12S = 0
16S = 2
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
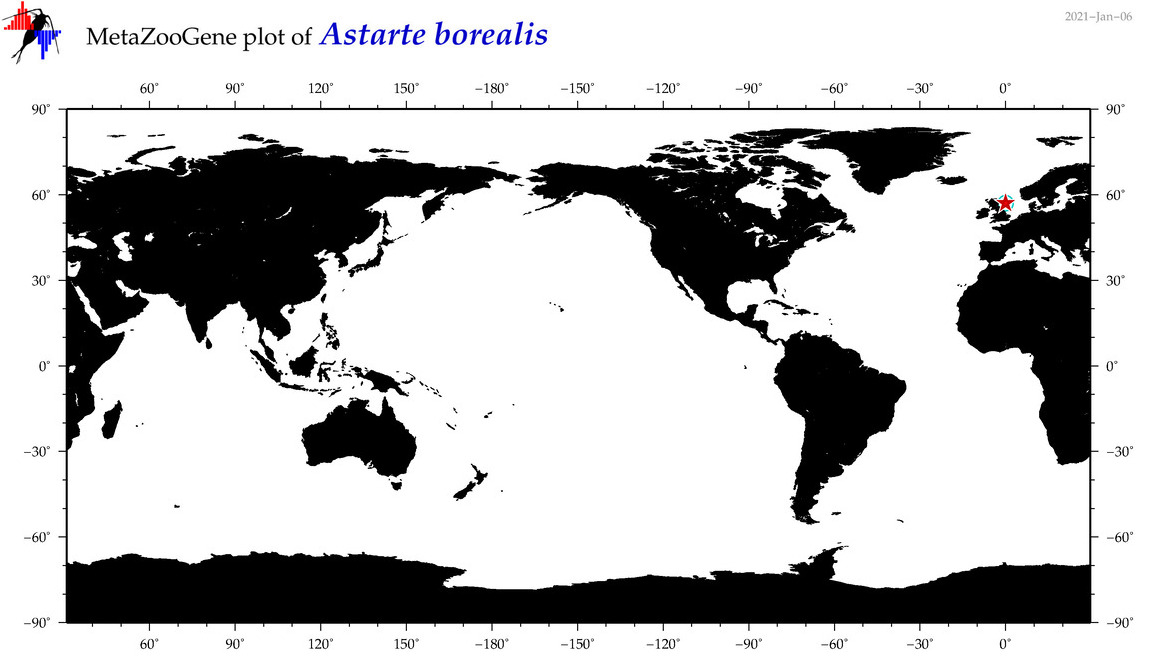 |
accepted
T4032720
R:1:0:0:0 |
| Astarte montagui |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 0
16S = 0
18S = 2
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
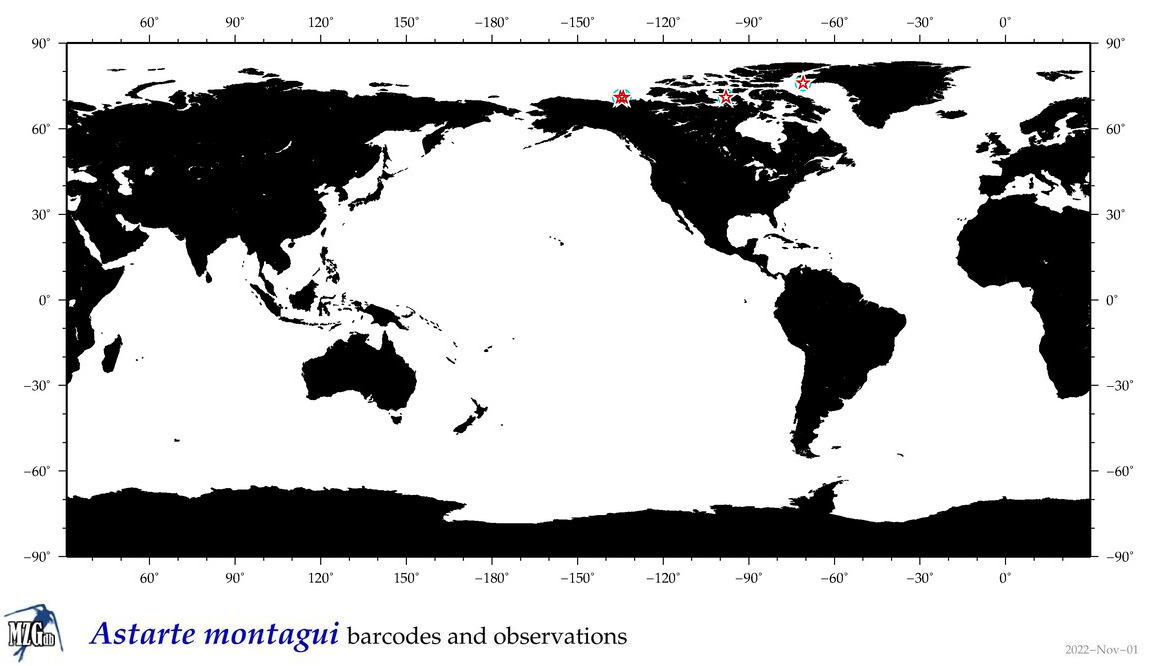 |
accepted
T4103716
R:1:0:0:0 |
| Astarte sulcata |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 1
18S = 2
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
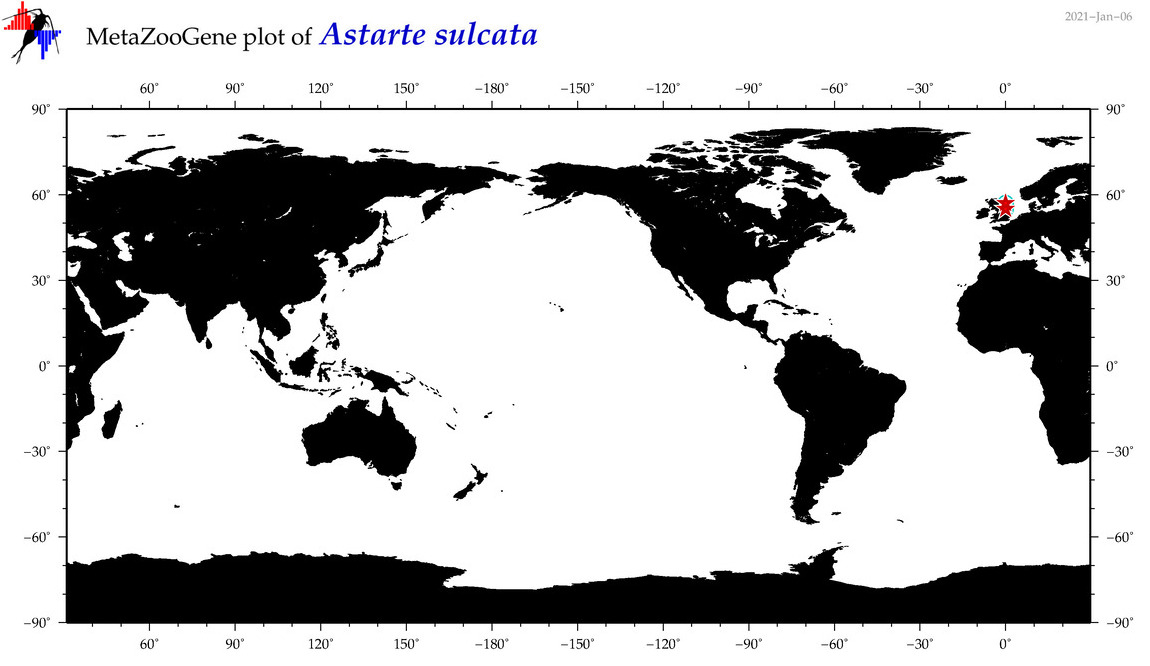 |
accepted
T4032721
R:1:0:0:0 |
| Barnea candida |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 1
18S = 3
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
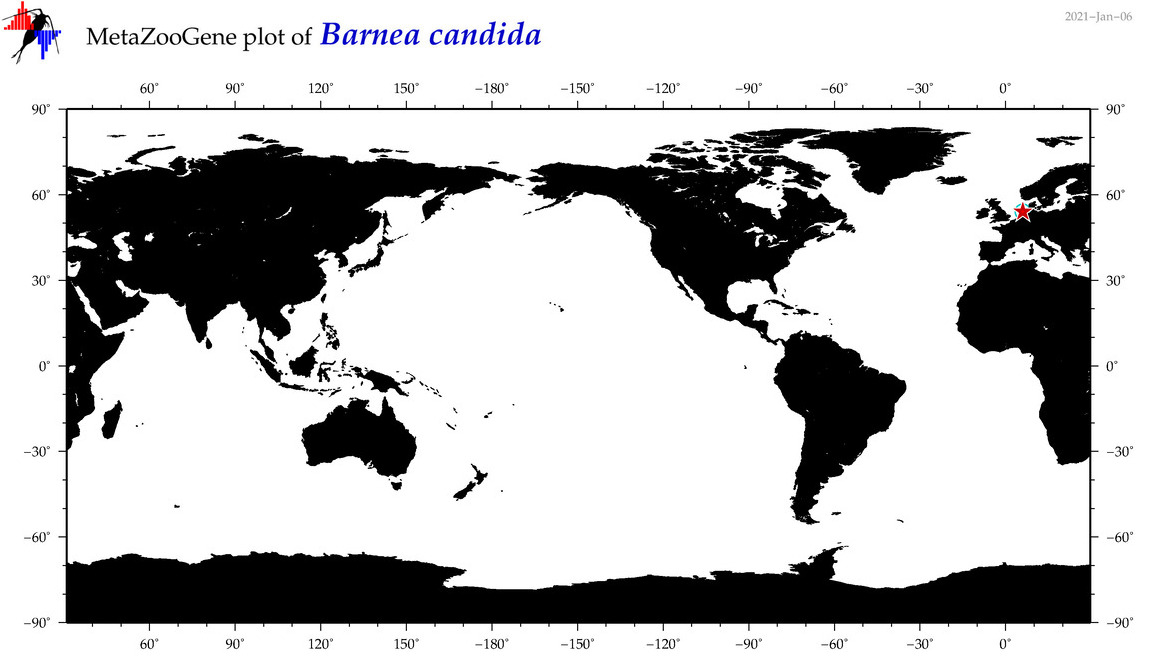 |
accepted
T4017079
R:1:0:0:0 |
| Cerastoderma edule |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 199
12S = 3
16S = 16
18S = 21
28S = 8
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
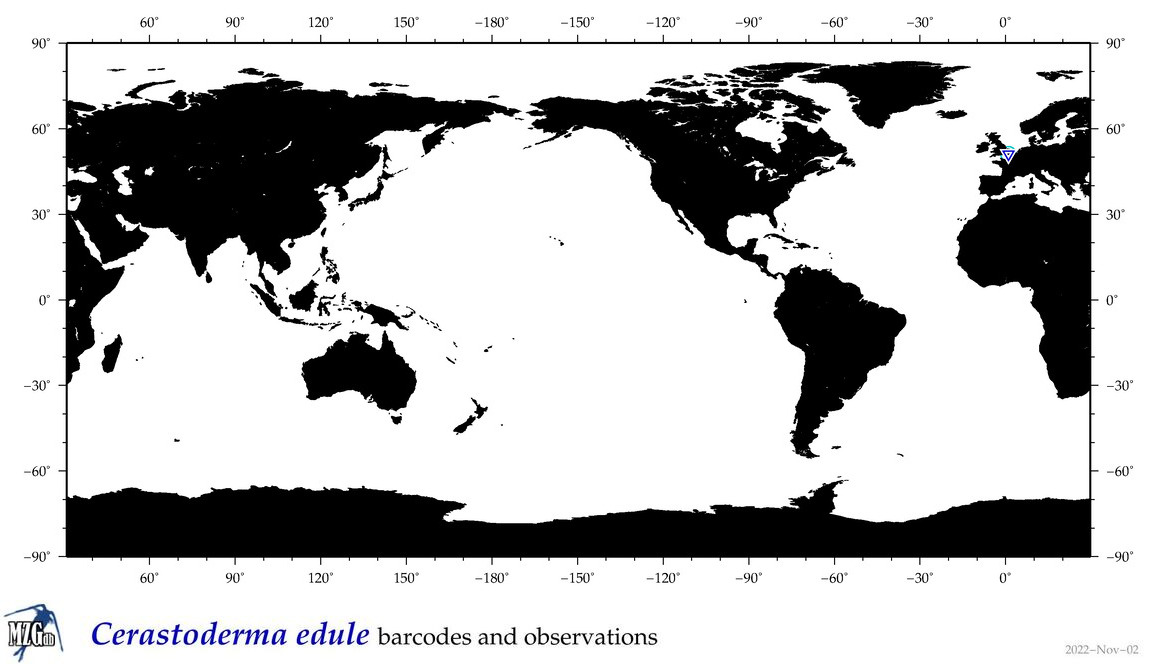 |
accepted
T4104025
R:1:0:0:0 |
| Chamelea gallina |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 87
12S = 0
16S = 21
18S = 3
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
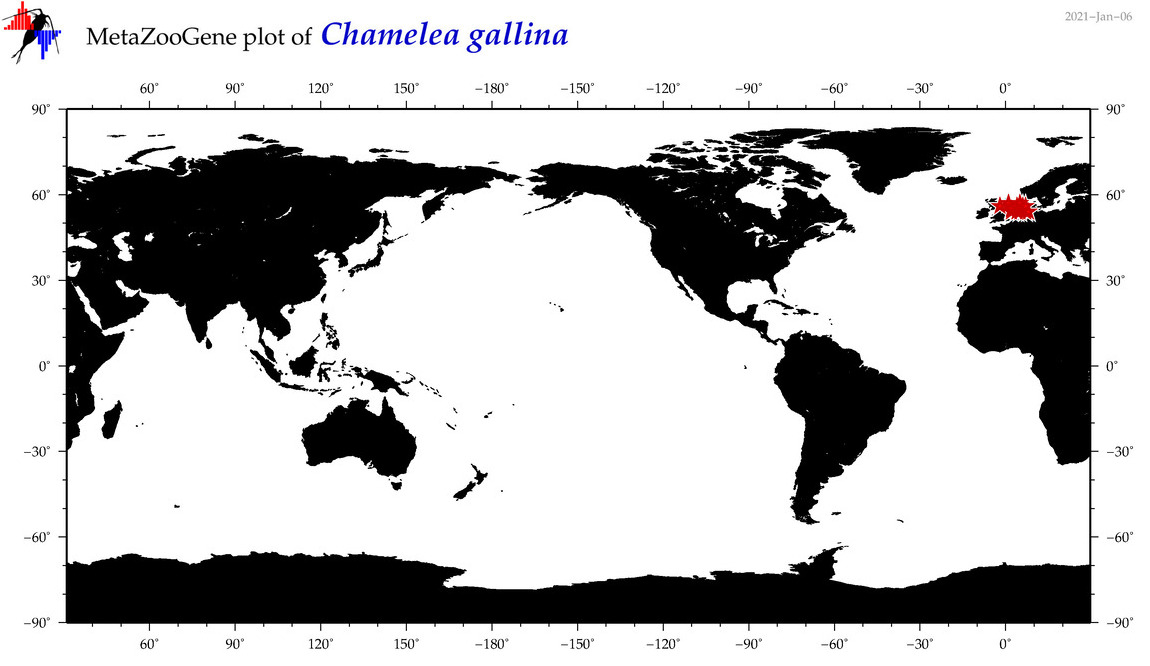 |
accepted
T4010754
R:1:0:0:0 |
| Chamelea striatula |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 0
16S = 7
18S = 1
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4104055
R:1:0:0:0 |
| Clausinella fasciata |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 2
18S = 2
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
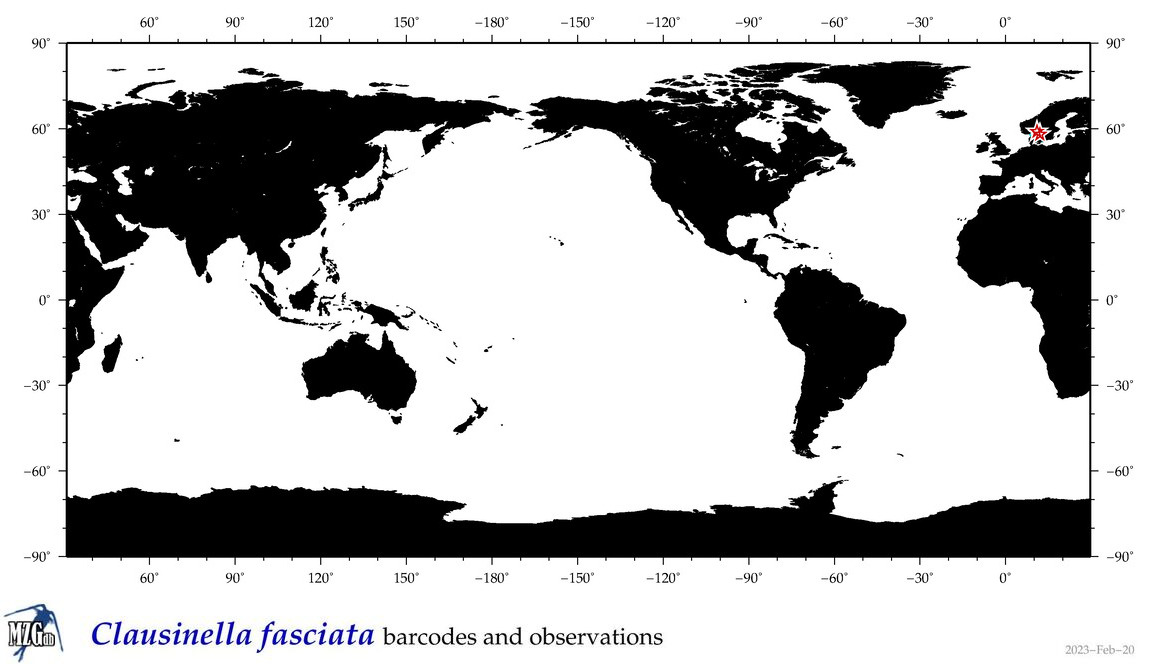 |
accepted
T4133955
R:1:0:0:0 |
| Cochlodesma praetenue |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 3
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
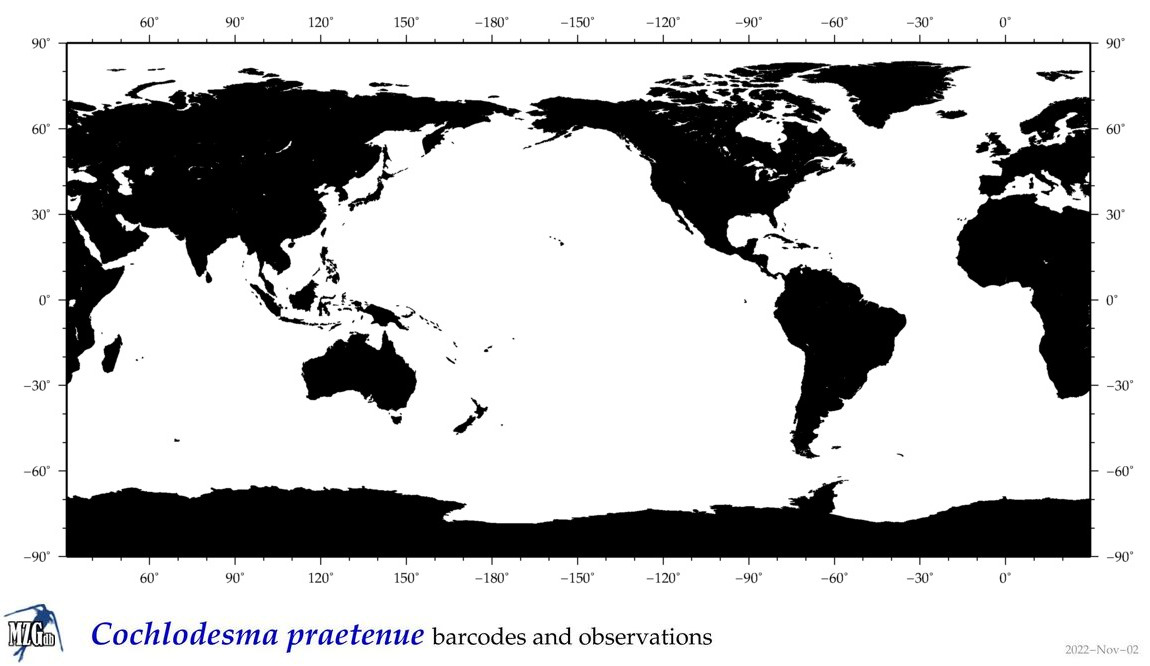 |
accepted
T4104155
R:1:0:0:0 |
| Cuspidaria cuspidata |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 4
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
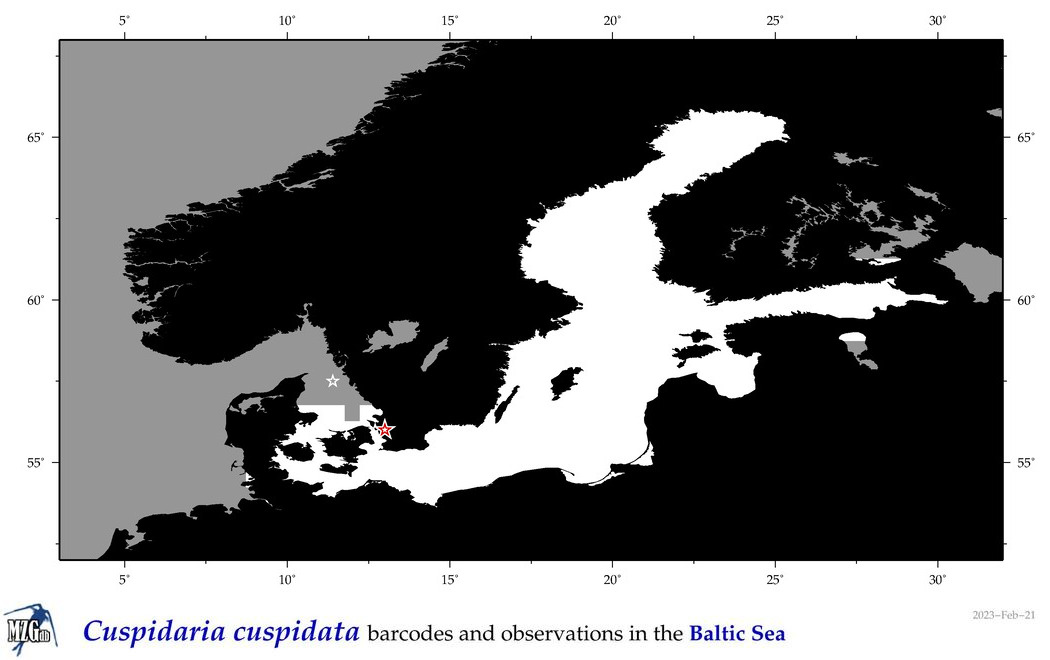 |
accepted
T4011055
R:1:0:0:0 |
| Dosinia exoleta |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 3
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
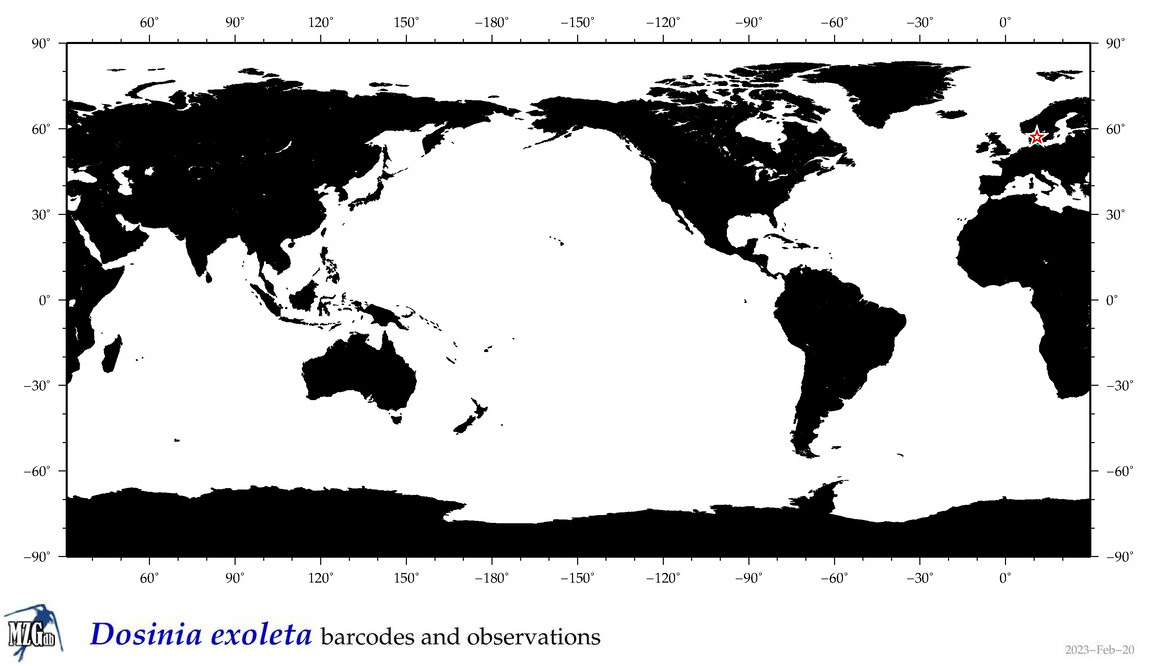 |
accepted
T4134353
R:1:0:0:0 |
| Dosinia lupinus |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
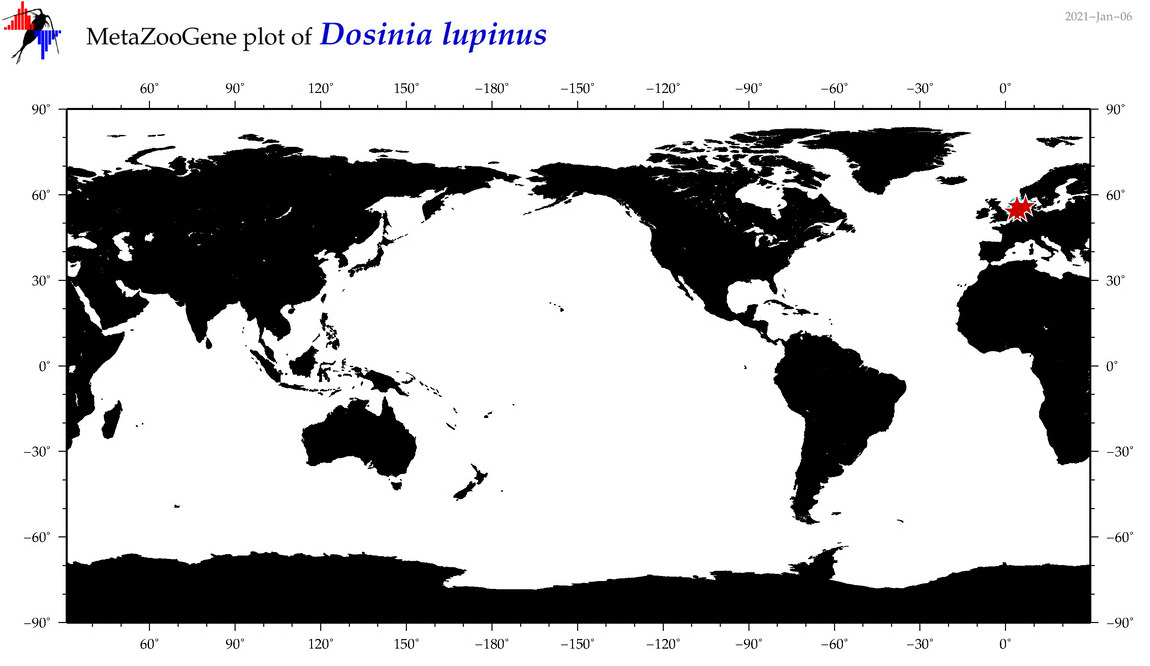 |
accepted
T4011295
R:1:0:0:0 |
| Dreissena polymorpha |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 117
12S = 3
16S = 20
18S = 5
28S = 14
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
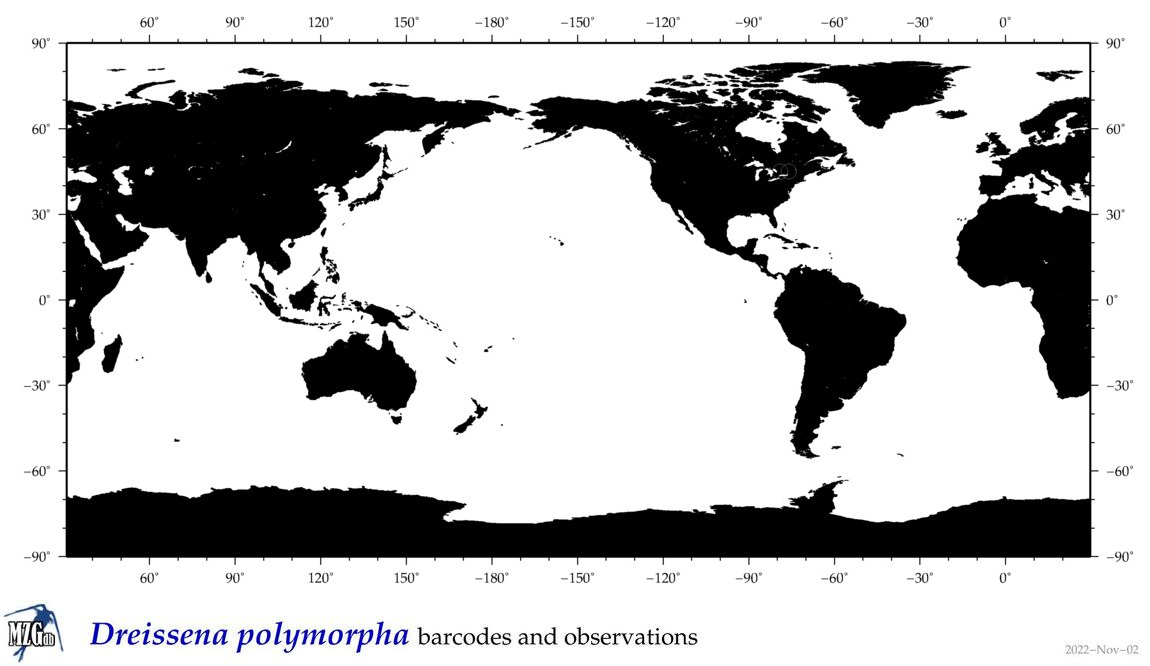 |
accepted
T4104439
R:0:1:1:0 |
| Ennucula tenuis |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 26
12S = 0
16S = 4
18S = 3
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
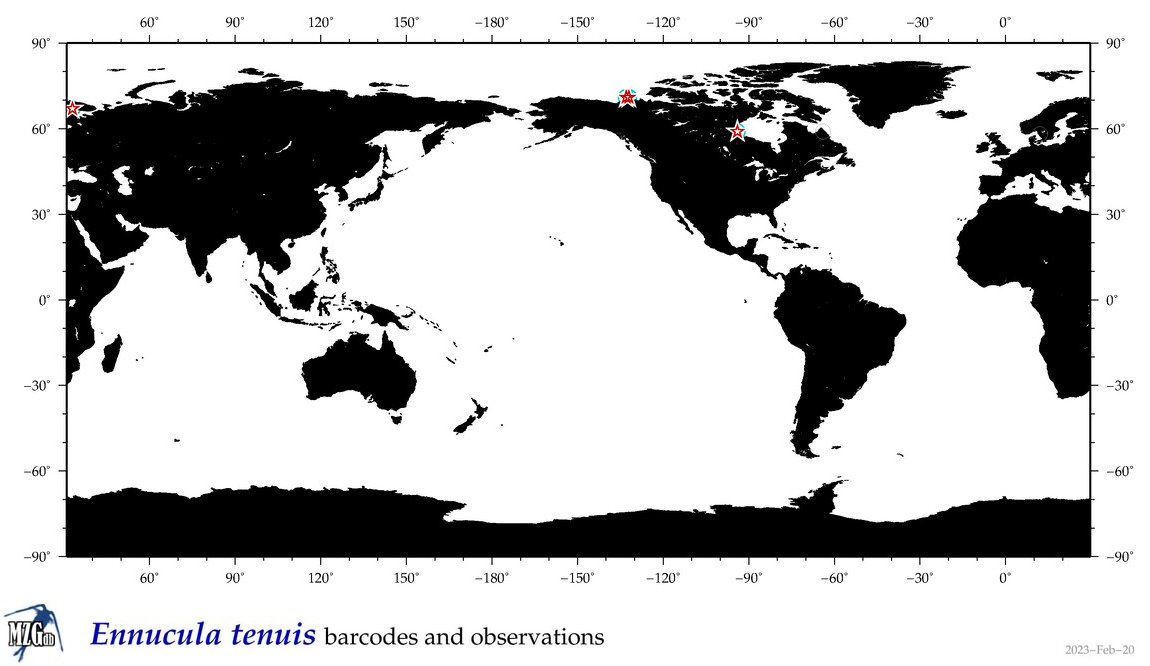 |
accepted
T4134501
R:1:0:0:0 |
| Ensis ensis |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 0
16S = 11
18S = 3
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
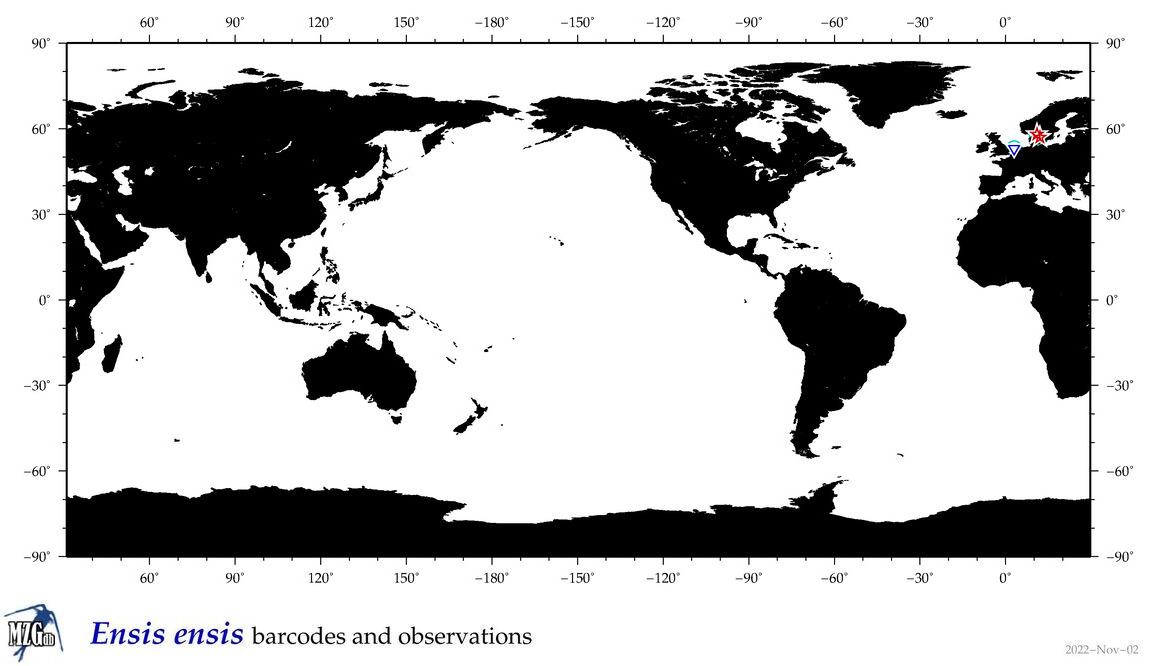 |
accepted
T4104539
R:1:0:0:0 |
| Ensis leei |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4134504
R:1:0:0:0 |
| Ensis siliqua |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 17
16S = 25
18S = 19
28S = 16
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
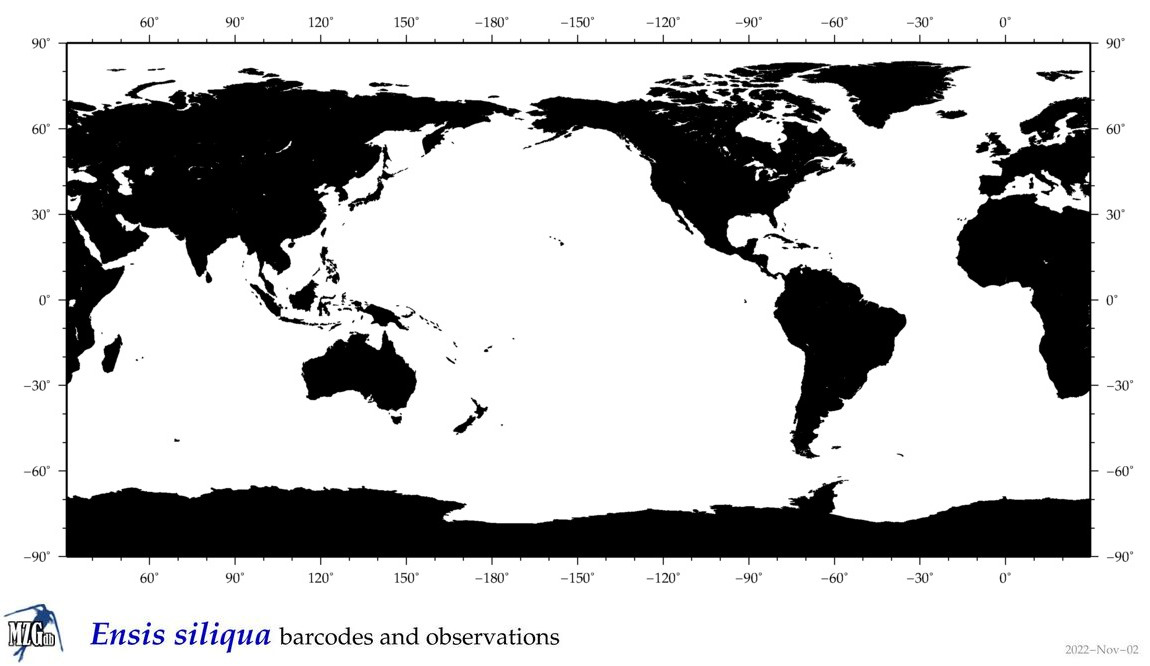 |
accepted
T4104542
R:1:0:0:0 |
| Gari fervensis |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
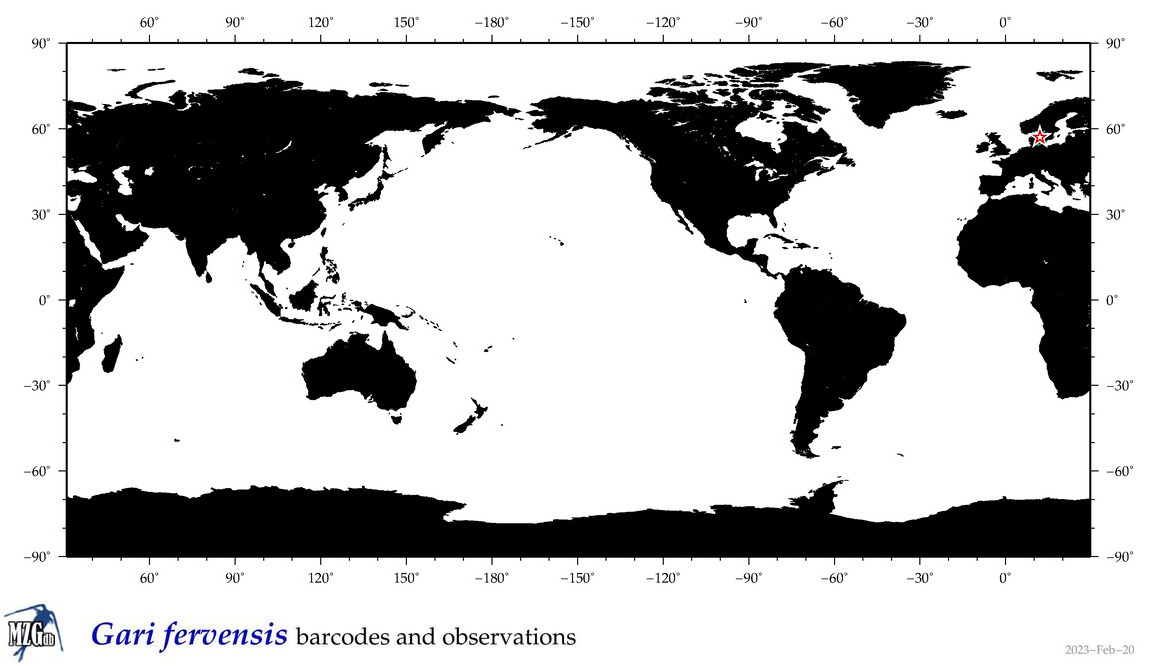 |
accepted
T4134792
R:1:0:0:0 |
| Hiatella arctica |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 11
18S = 3
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4003123
R:1:0:0:0 |
| Kurtiella bidentata |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
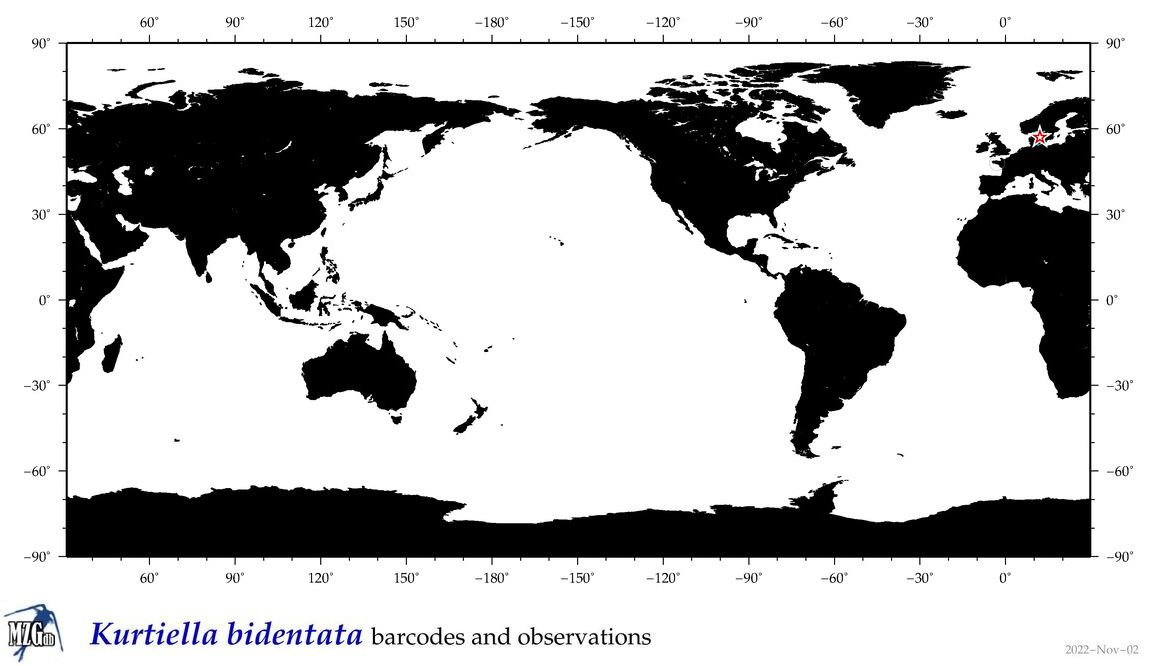 |
accepted
T4105148
R:1:0:0:0 |
| Laevicardium crassum |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 3
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
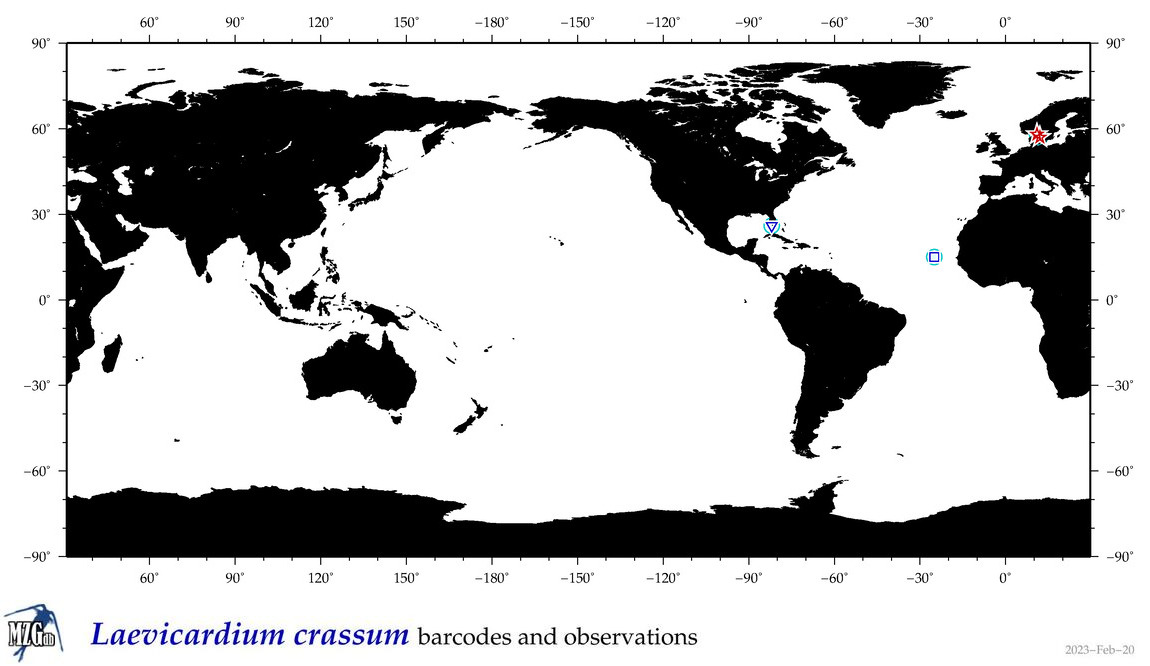 |
accepted
T4135176
R:1:0:0:0 |
| Limecola balthica |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 848
12S = 4
16S = 12
18S = 16
28S = 2
|
COI = 13
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4017088
R:1:0:0:0 |
| Lucinoma borealis |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 5
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
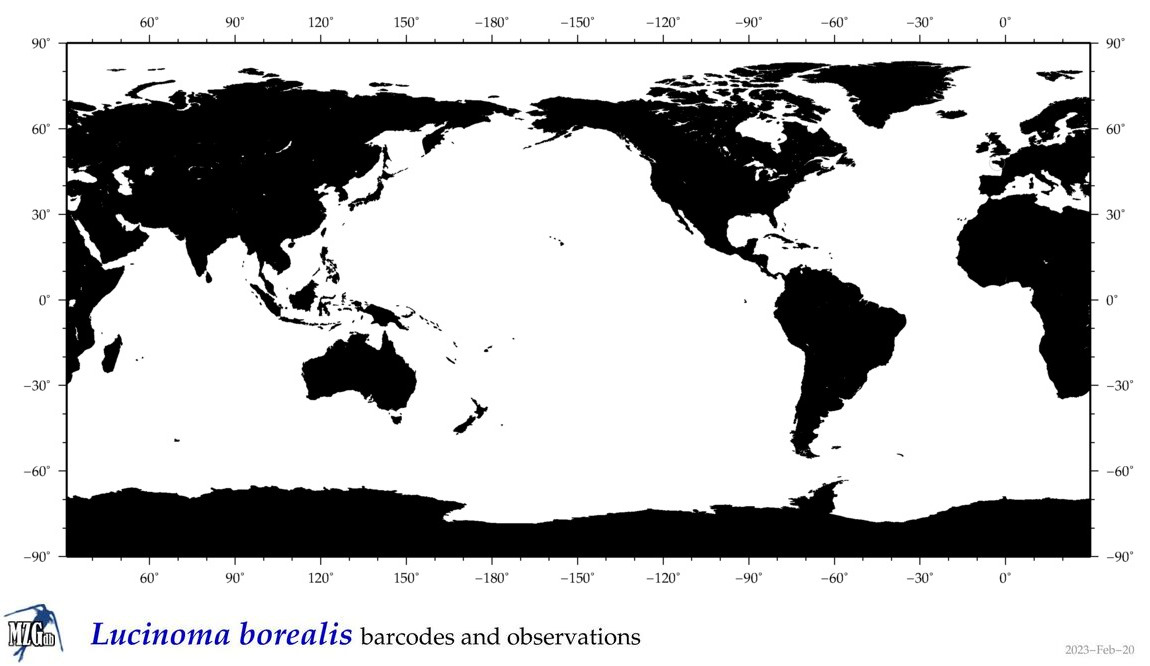 |
accepted
T4105303
R:1:0:0:0 |
| Macoma calcarea |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 124
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 17
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4025464
R:1:0:0:0 |
| Macomangulus tenuis |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 3
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
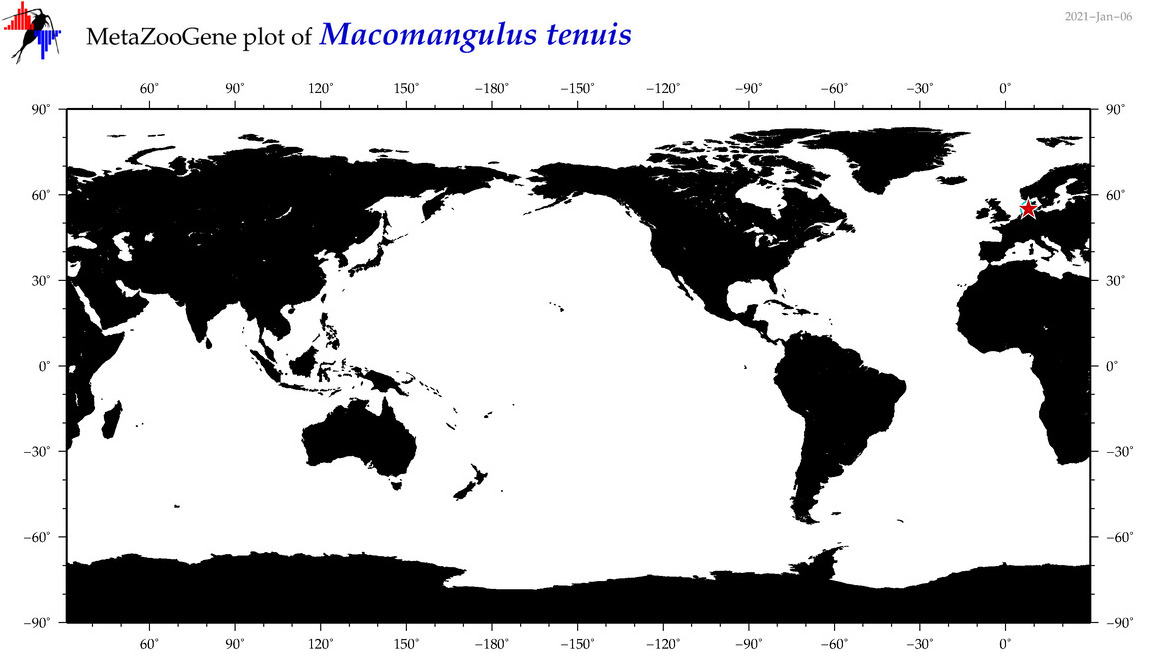 |
accepted
T4098408
R:1:0:0:0 |
| Magallana gigas |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 650
12S = 1
16S = 57
18S = 23
28S = 30
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4098403
R:1:1:0:0 |
| Modiolula phaseolina |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
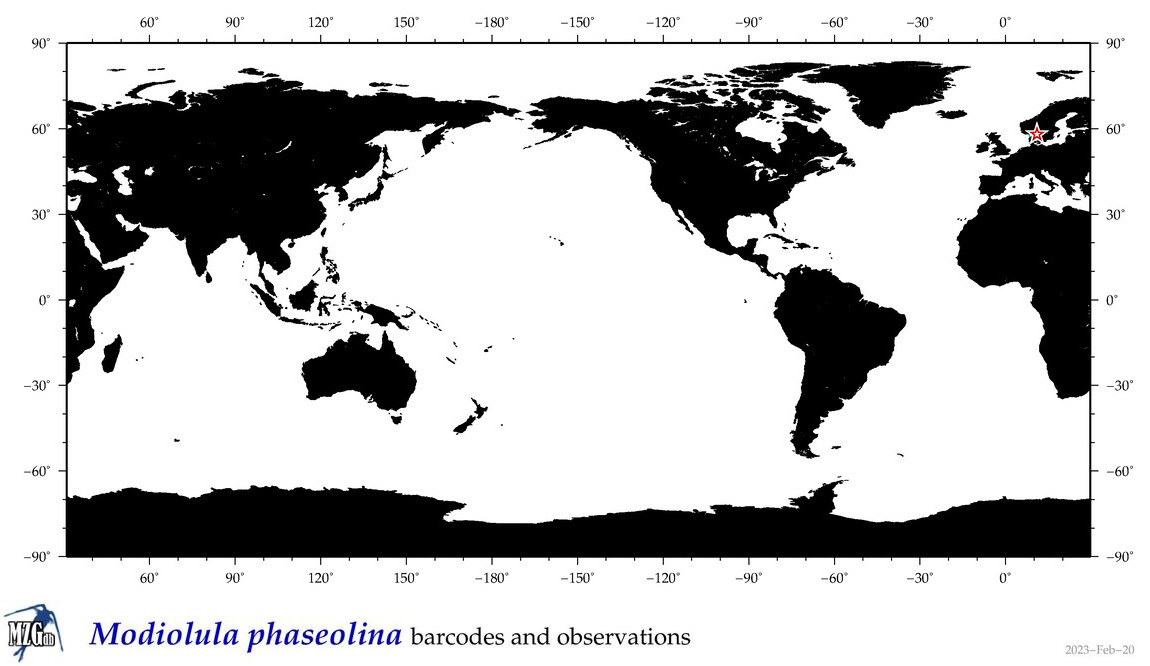 |
accepted
T4135559
R:1:0:0:0 |
| Modiolus modiolus |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 287
12S = 3
16S = 16
18S = 4
28S = 5
|
COI = 26
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4013089
R:1:0:0:0 |
| Musculus discors |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 0
16S = 7
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
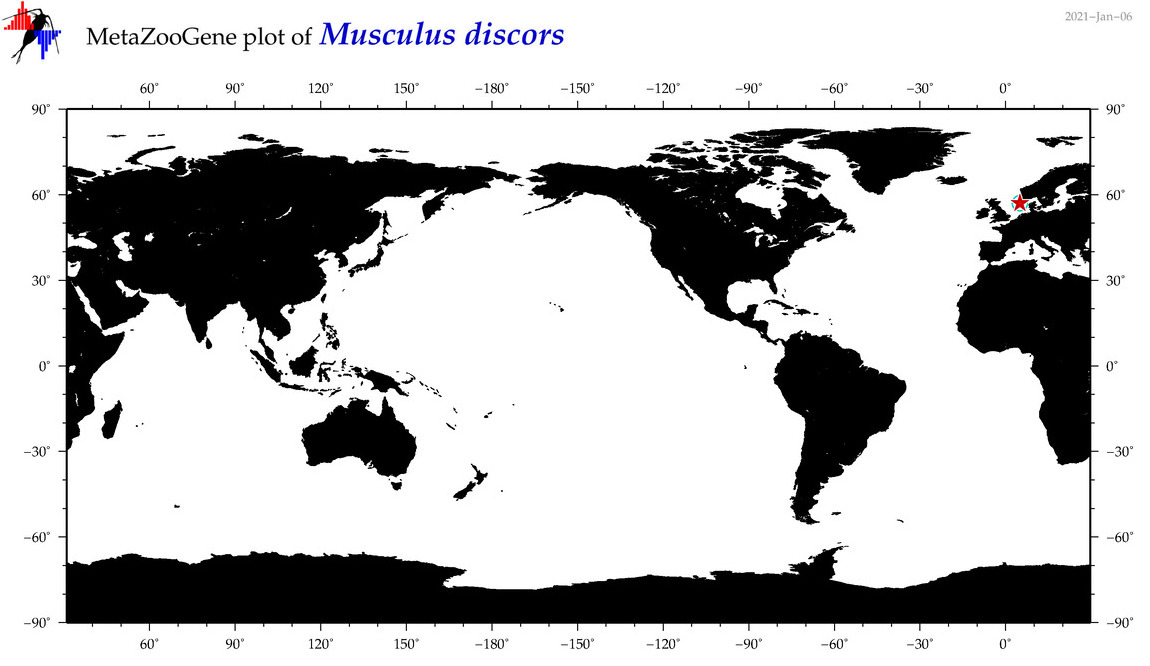 |
accepted
T4017177
R:1:0:0:0 |
| Musculus niger |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
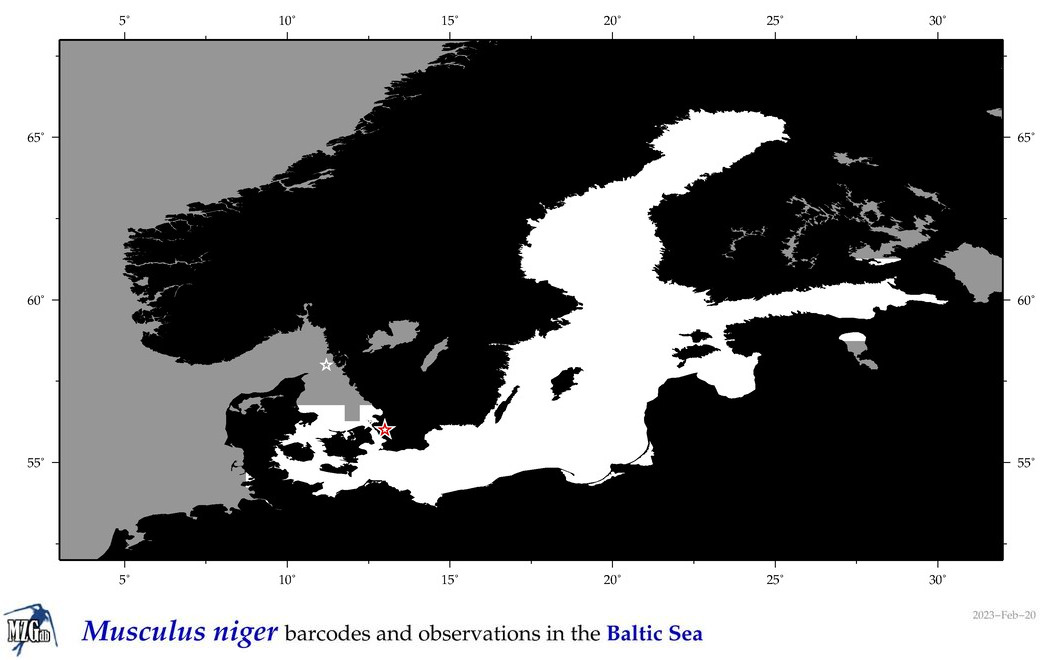 |
accepted
T4105511
R:1:0:0:0 |
| Mya arenaria |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 138
12S = 3
16S = 25
18S = 7
28S = 6
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
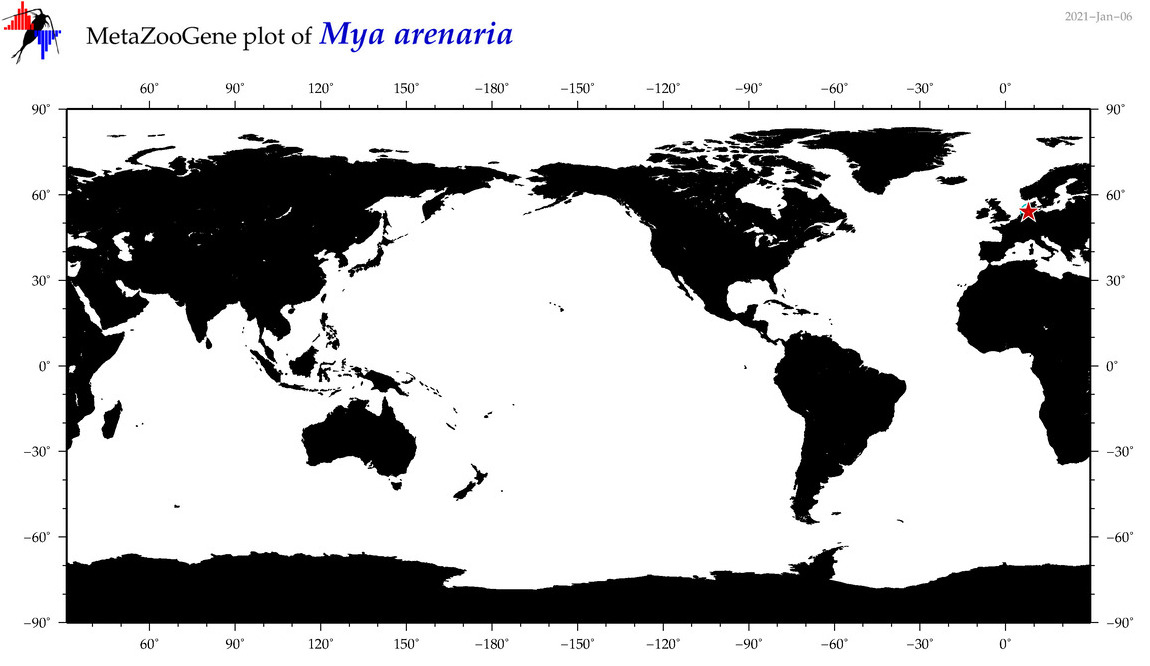 |
accepted
T4022213
R:1:0:0:0 |
| Mya truncata |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 0
16S = 2
18S = 3
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
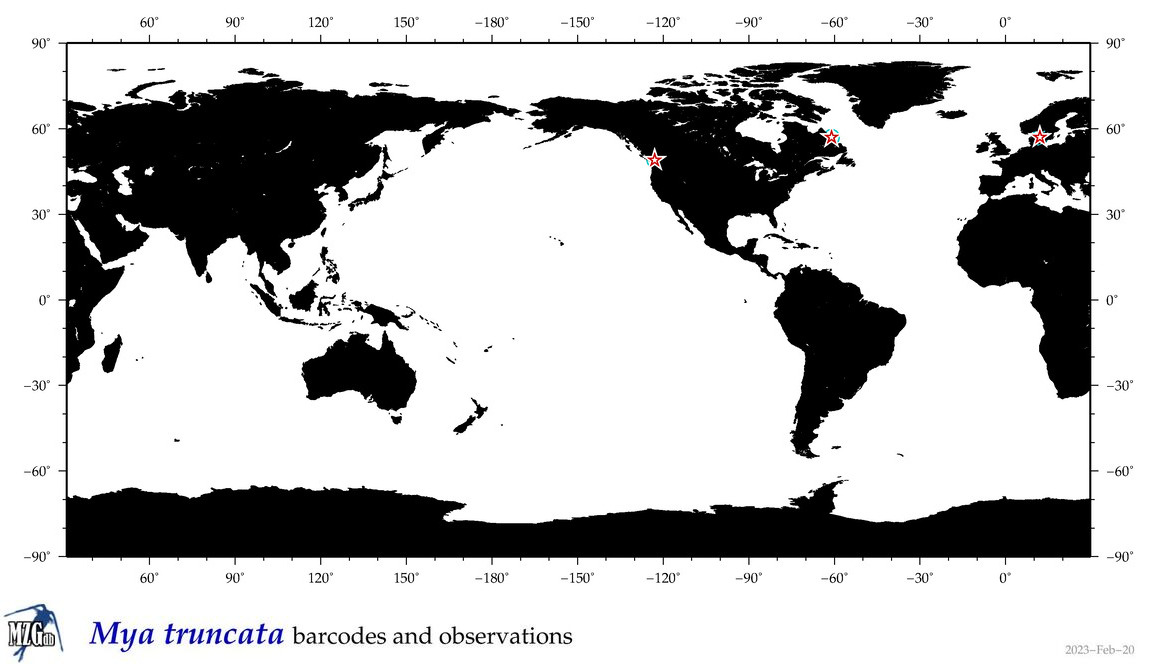 |
accepted
T4105516
R:1:0:0:0 |
| Mysia undata |
Species
(41) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
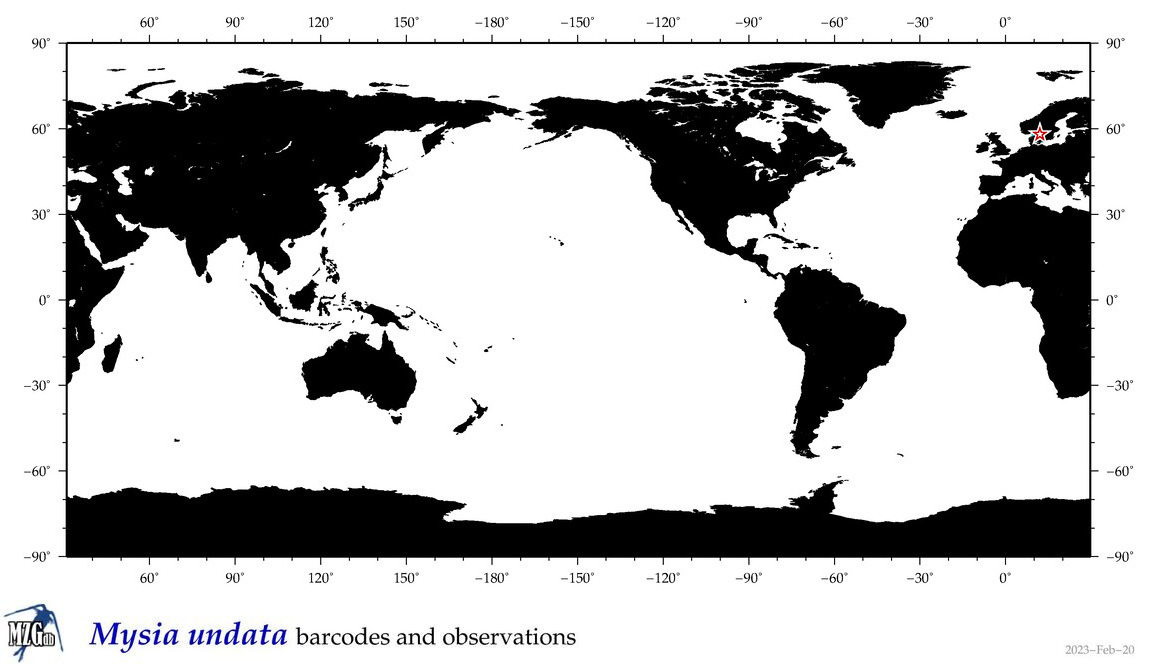 |
accepted
T4105540
R:1:0:0:0 |
| Mytilopsis leucophaeata |
Species
(42) |
COI
12S
16S
18S
28S
|
COI = 143
12S = 1
16S = 7
18S = 2
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4105543
R:0:1:1:0 |
| Mytilus edulis |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 705
12S = 5
16S = 639
18S = 10
28S = 7
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
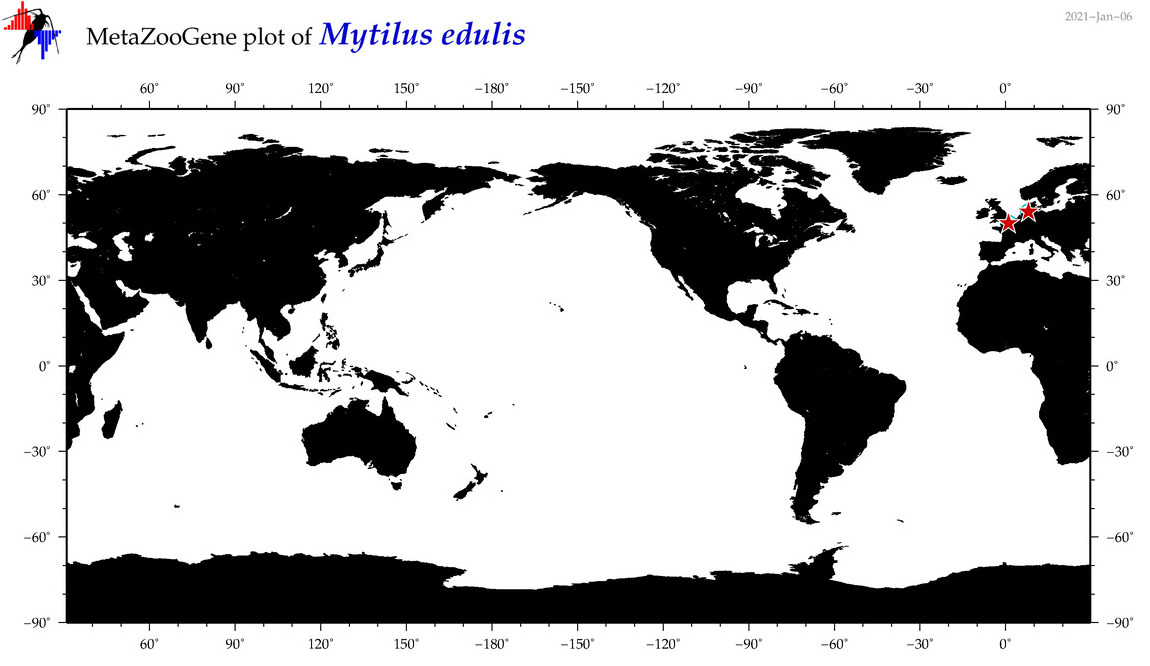 |
accepted
T4000445
R:1:0:0:0 |
| Mytilus trossulus |
Species
(44) |
COI
12S
16S
18S
28S
|
COI = 1009
12S = 14
16S = 311
18S = 8
28S = 3
|
COI = 0
12S = 0
16S = 155
18S = 0
28S = 0
|
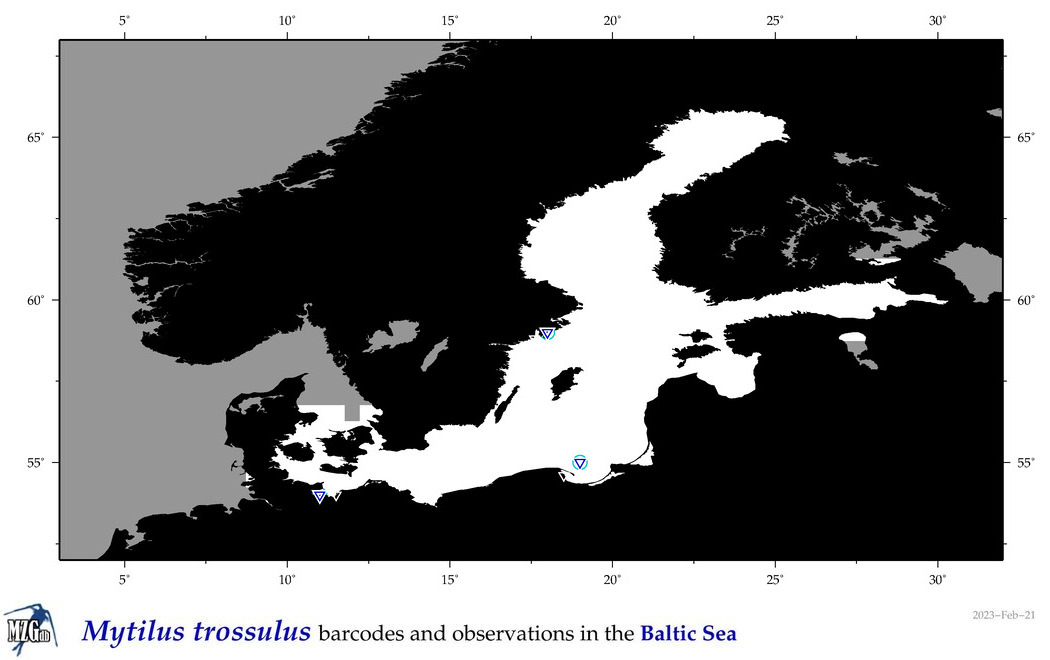 |
accepted
T4000839
R:1:0:0:0 |
| Nuculana minuta |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 2
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
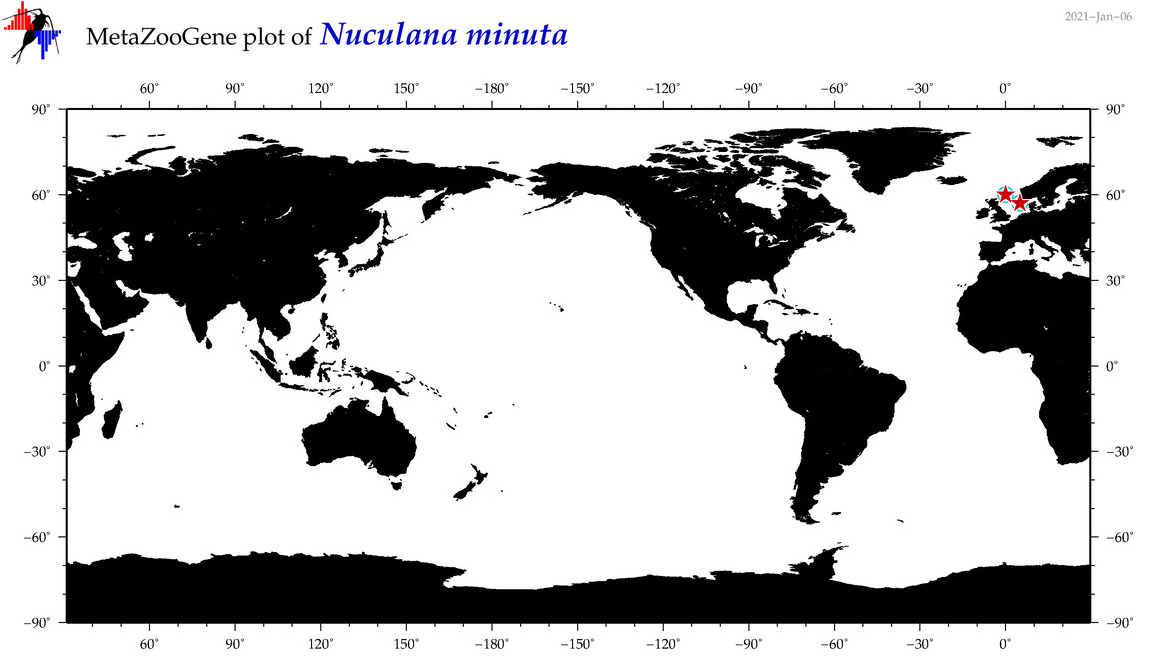 |
accepted
T4013426
R:1:0:0:0 |
| Nuculana pernula |
Species
(46) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 0
16S = 0
18S = 3
28S = 3
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
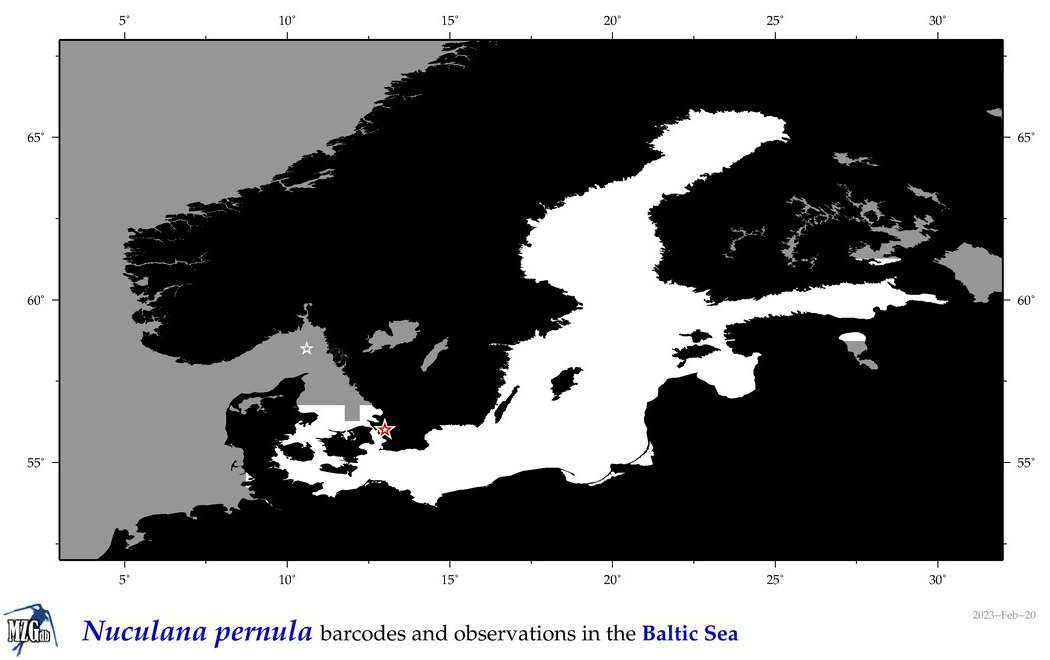 |
accepted
T4105689
R:1:0:0:0 |
| Nucula nitidosa |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 23
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
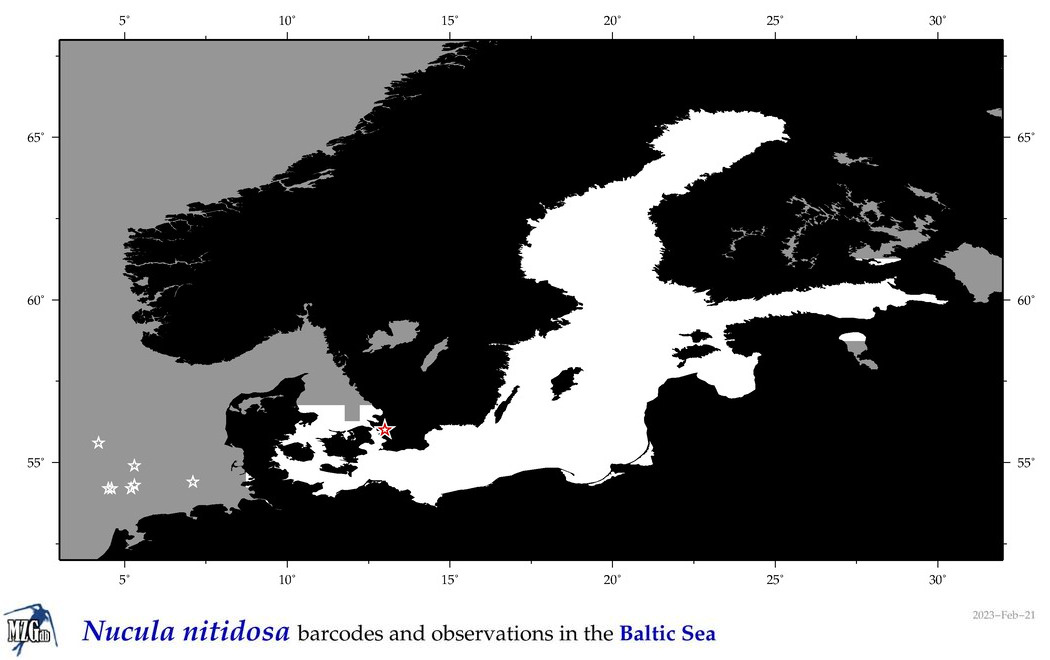 |
accepted
T4022245
R:1:0:0:0 |
| Nucula nucleus |
Species
(48) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 1
16S = 1
18S = 8
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4135705
R:1:0:0:0 |
| Parathyasira equalis |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 19
12S = 0
16S = 1
18S = 3
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
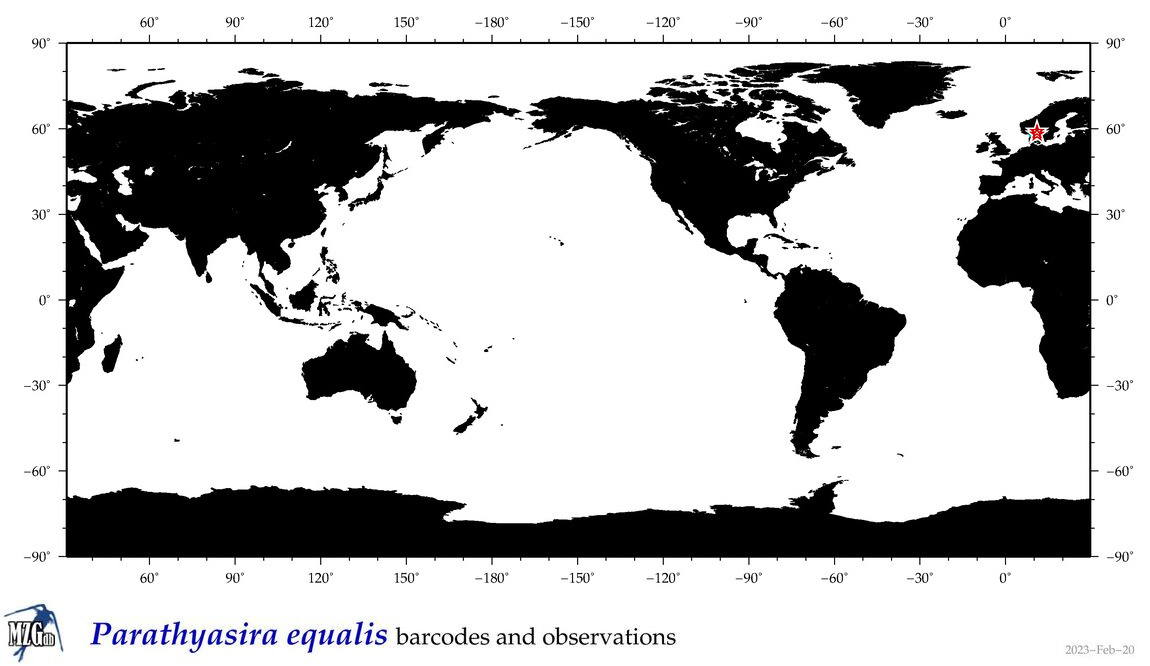 |
accepted
T4105885
R:1:0:0:0 |
| Parvicardium exiguum |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4105909
R:1:0:0:0 |
| Parvicardium pinnulatum |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
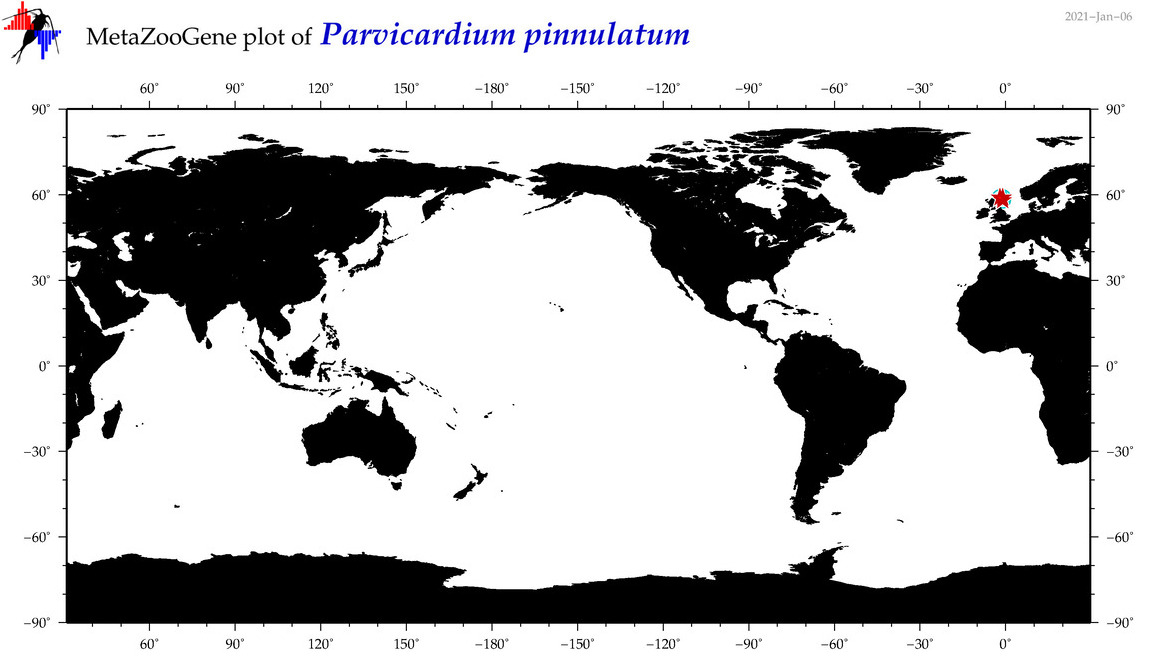 |
accepted
T4017096
R:1:0:0:0 |
| Parvicardium scabrum |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
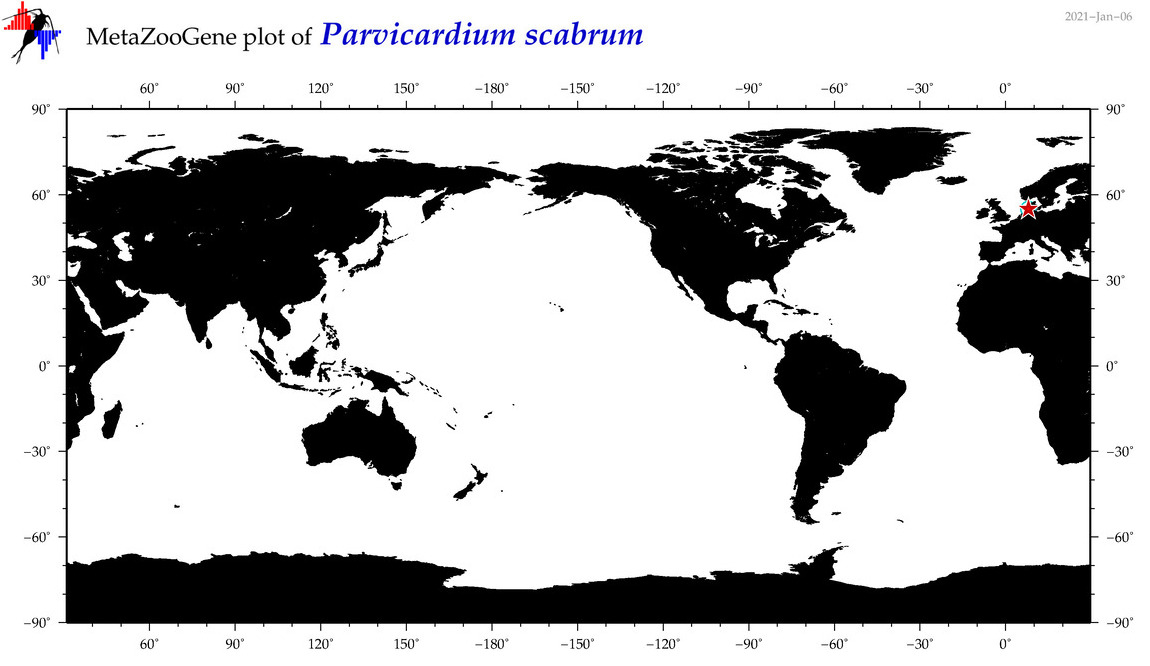 |
accepted
T4017097
R:1:0:0:0 |
| Petricolaria pholadiformis |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 6
28S = 4
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
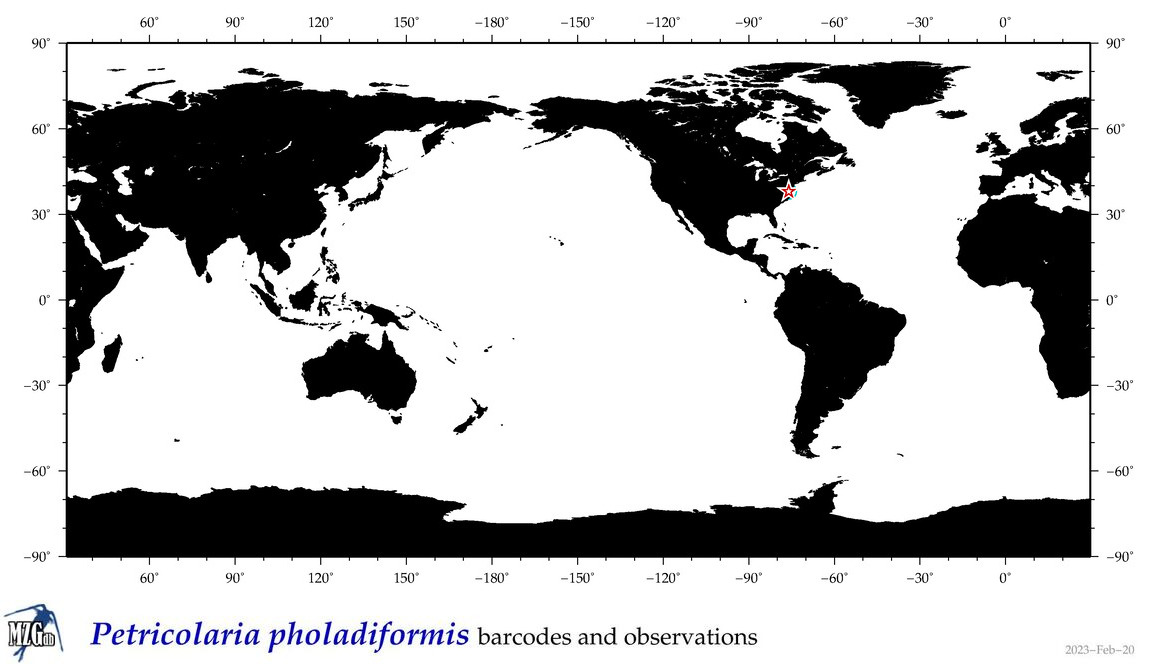 |
accepted
T4105945
R:1:0:0:0 |
| Phaxas pellucidus |
Species
(54) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 0
16S = 2
18S = 4
28S = 2
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4016251
R:1:0:0:0 |
| Pododesmus patelliformis |
Species
(55) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4092029
R:1:0:0:0 |
| Rangia cuneata |
Species
(56) |
COI
12S
16S
18S
28S
|
COI = 22
12S = 0
16S = 5
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
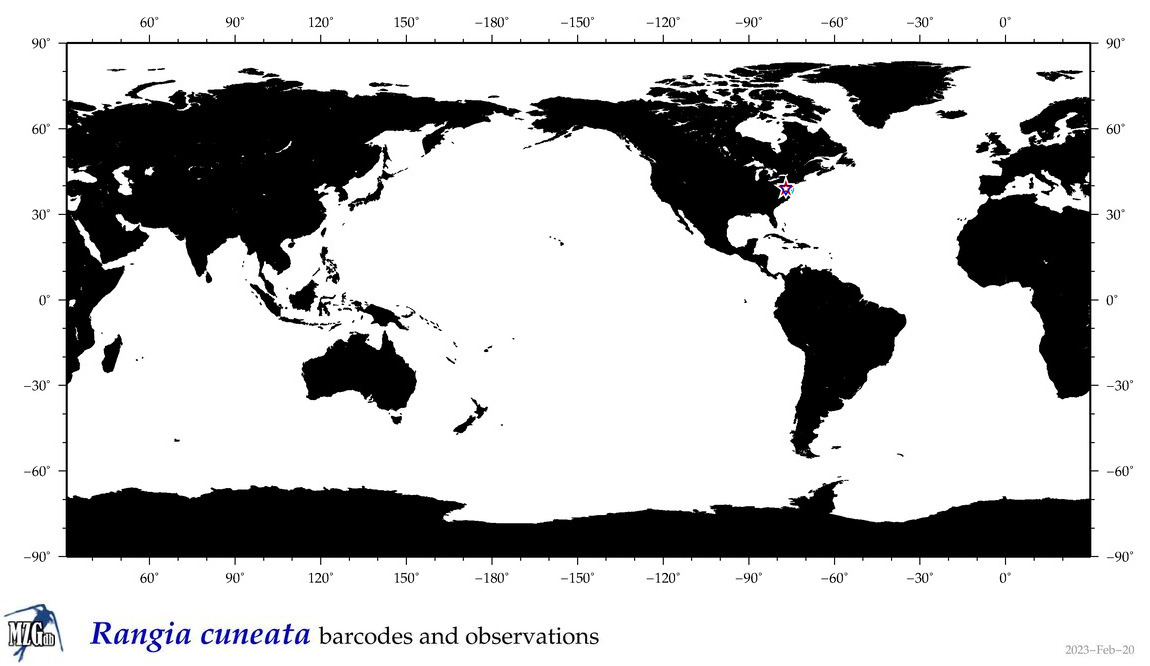 |
accepted
T4106300
R:0:1:1:0 |
| Scrobicularia plana |
Species
(57) |
COI
12S
16S
18S
28S
|
COI = 74
12S = 1
16S = 2
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4106421
R:1:0:0:0 |
| Spisula elliptica |
Species
(58) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 0
16S = 0
18S = 2
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4015043
R:1:0:0:0 |
| Spisula solida |
Species
(59) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 3
18S = 6
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4015044
R:1:0:0:0 |
| Spisula subtruncata |
Species
(60) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
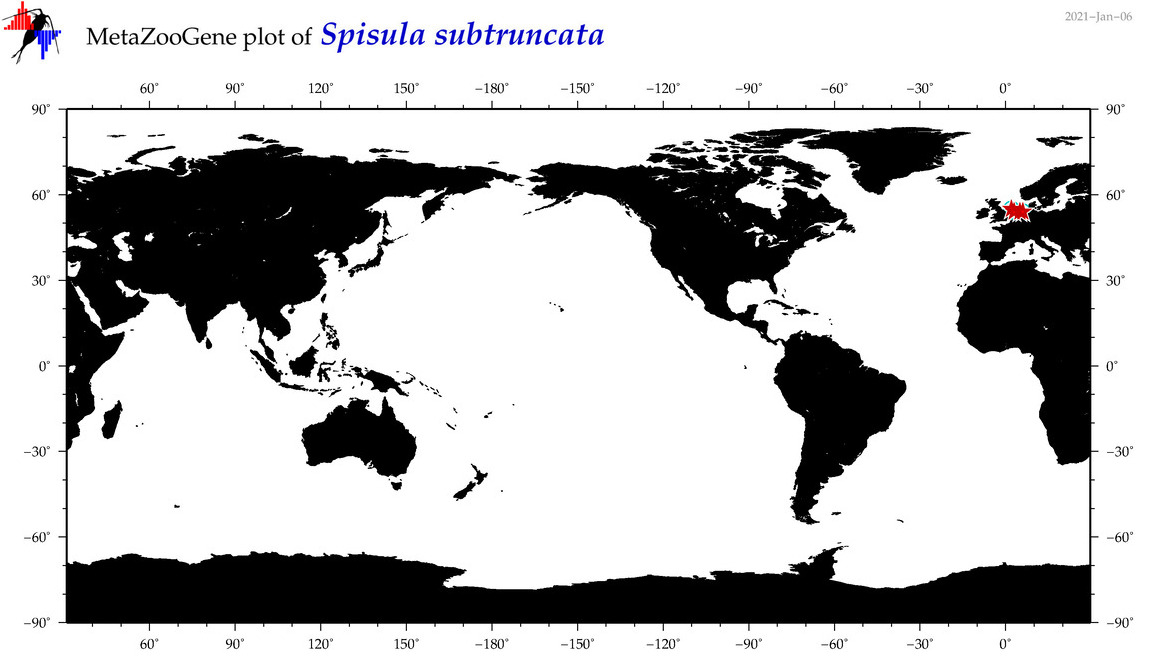 |
accepted
T4015045
R:1:0:0:0 |
| Tellimya ferruginosa |
Species
(61) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 1
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4106626
R:1:0:0:0 |
| Teredo navalis |
Species
(62) |
COI
12S
16S
18S
28S
|
COI = 158
12S = 0
16S = 0
18S = 4
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4136108
R:1:1:0:0 |
| Thracia phaseolina |
Species
(63) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 1
18S = 1
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
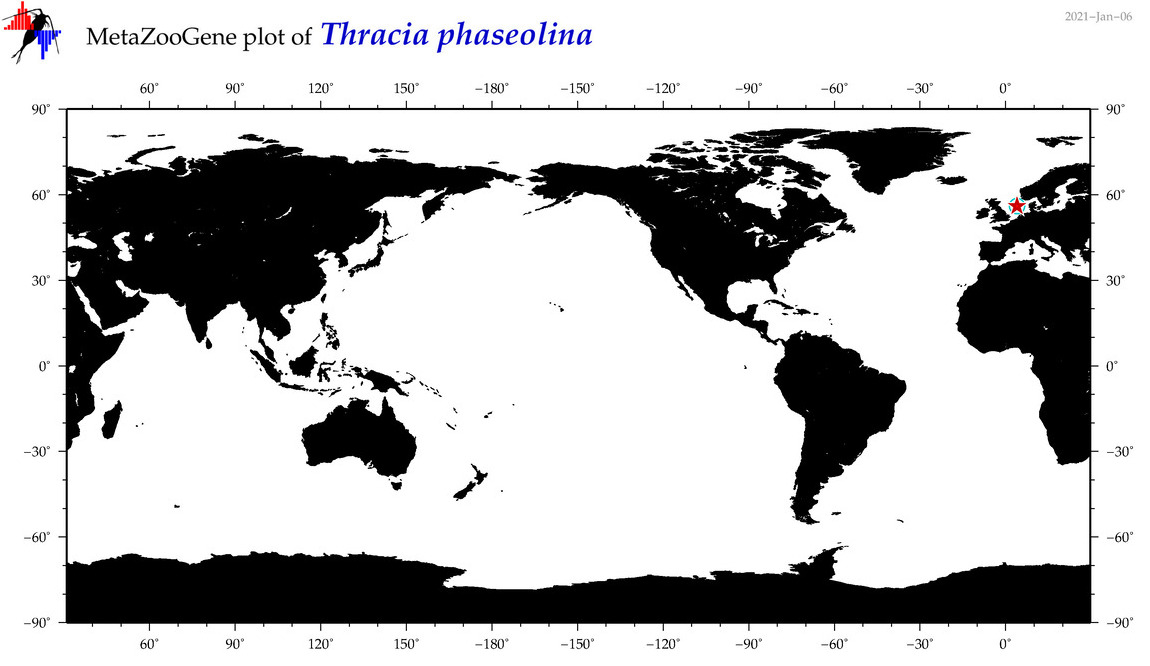 |
accepted
T4015375
R:1:0:0:0 |
| Thyasira flexuosa |
Species
(64) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 0
18S = 5
28S = 4
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
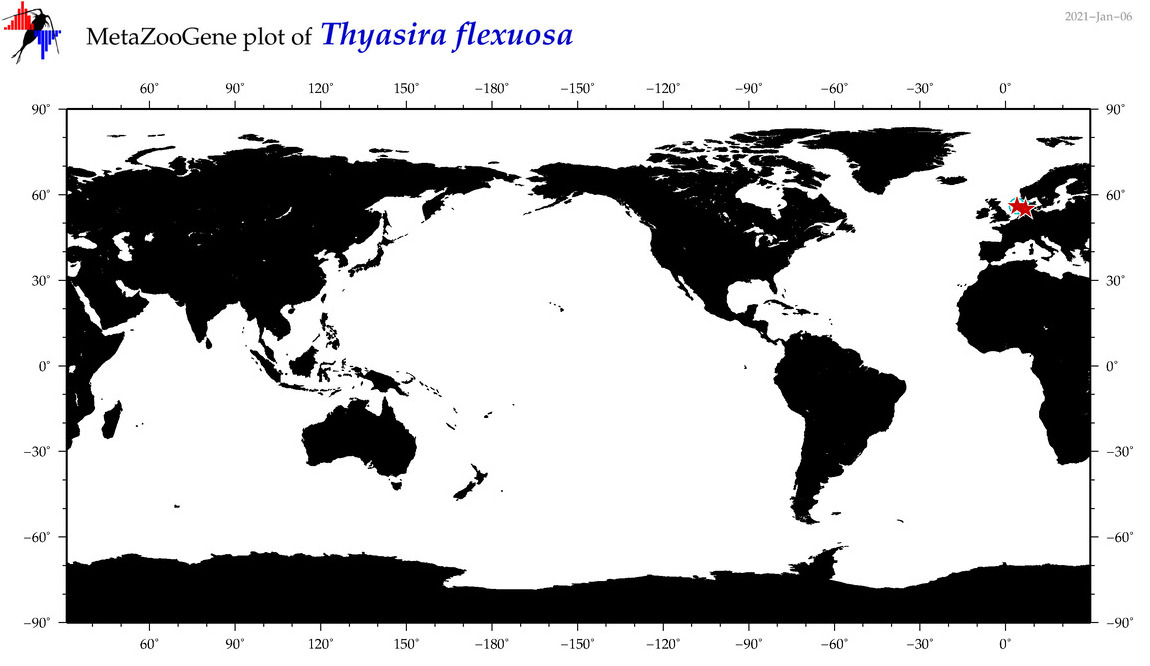 |
accepted
T4016332
R:1:0:0:0 |
| Timoclea ovata |
Species
(65) |
COI
12S
16S
18S
28S
|
COI = 19
12S = 1
16S = 1
18S = 4
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
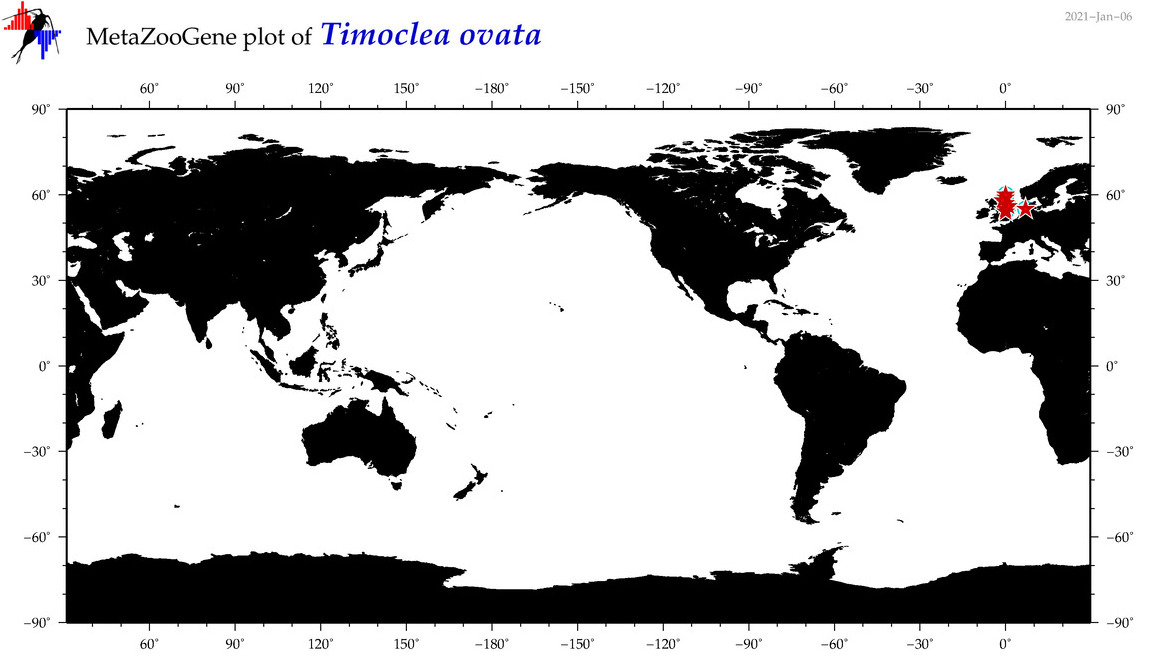 |
accepted
T4015396
R:1:0:0:0 |
| Varicorbula gibba |
Species
(66) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 13
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094076
R:1:0:0:0 |
| Venus casina |
Species
(67) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
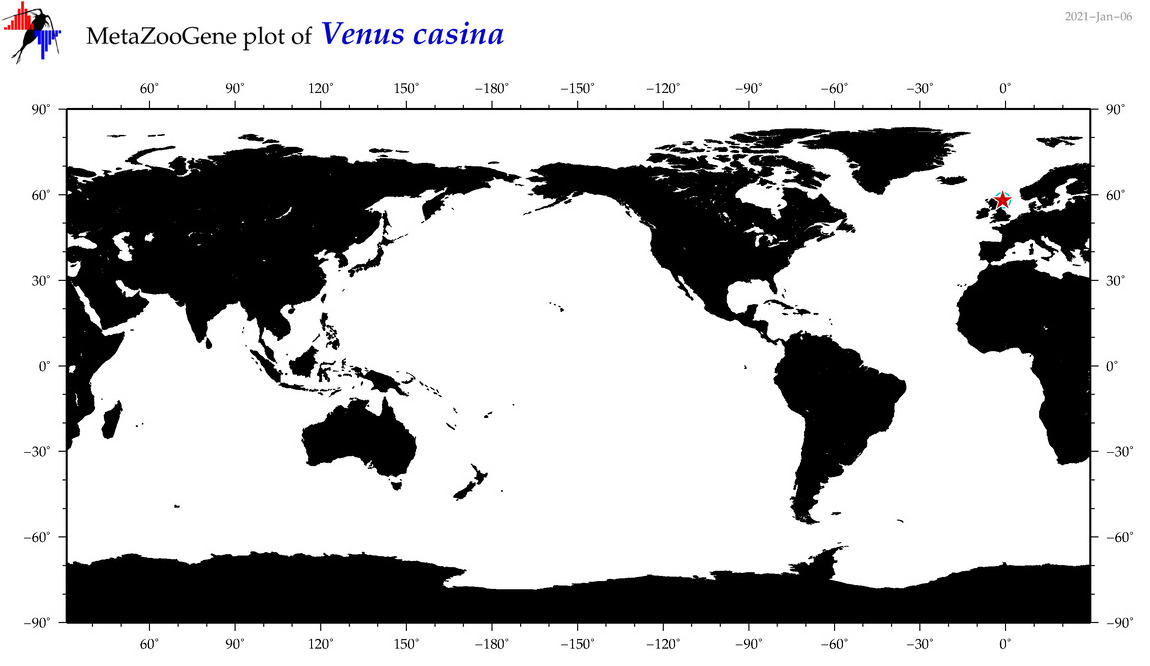 |
accepted
T4015566
R:1:0:0:0 |