Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2023-m07-15
2023-Jul-20 |
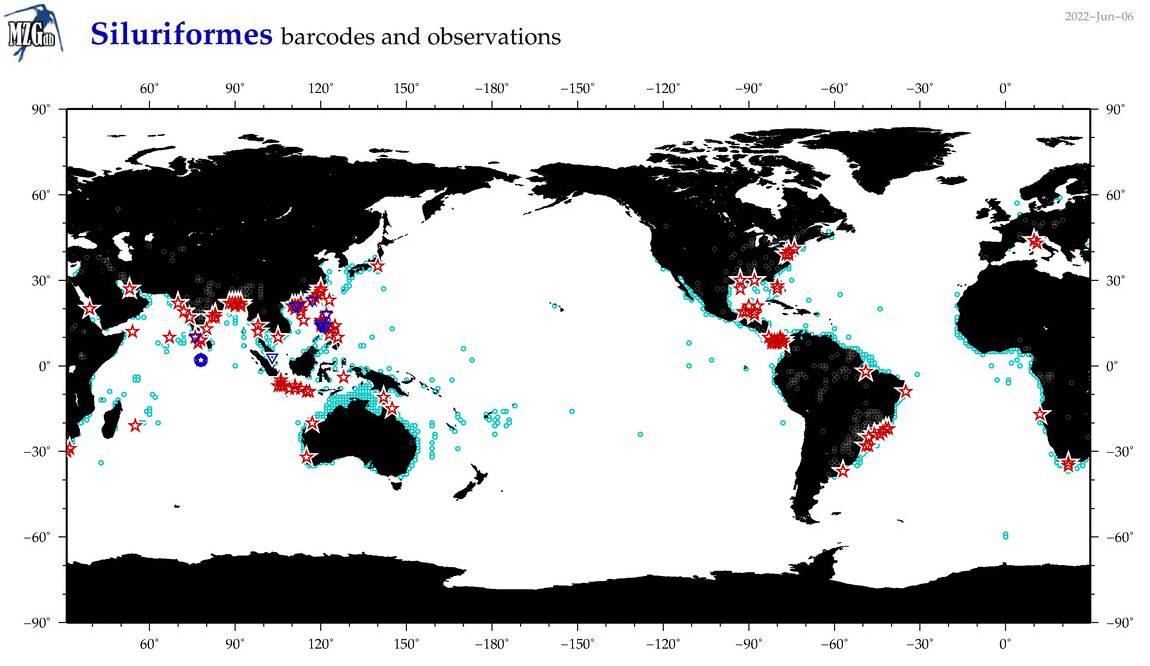
| Siluriformes |
Order |
cummulative
sub-totals
------->
for this
Order |
Total Species: 186
Sp. w/COI-Barcodes: 112
Total COI-Barcodes: 3052 |
Total Species: 186
Sp. w/COI-Barcodes: 112
Total COI-Barcodes: 3052 |
 |
accepted
T5002222
R:1:1:1:0 |
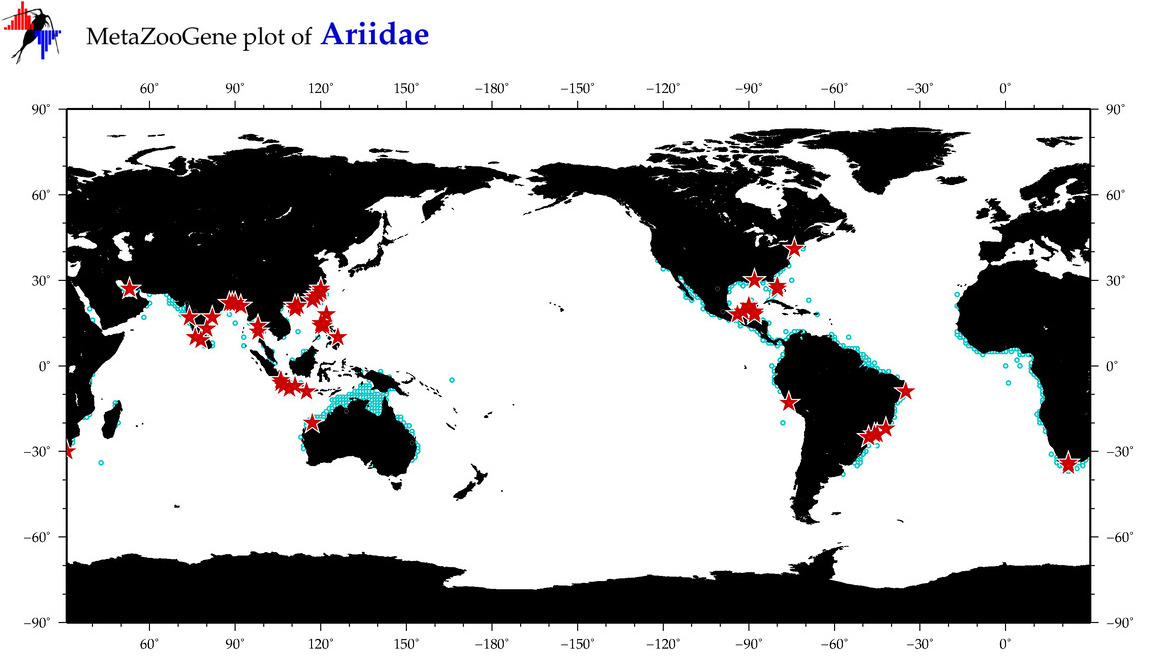
| ⋄ Ariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 100
Sp. w/COI-Barcodes: 41
Total COI-Barcodes: 696 |
Total Species: 100
Sp. w/COI-Barcodes: 41
Total COI-Barcodes: 696 |
 |
accepted
T5006029
R:1:1:1:0 |
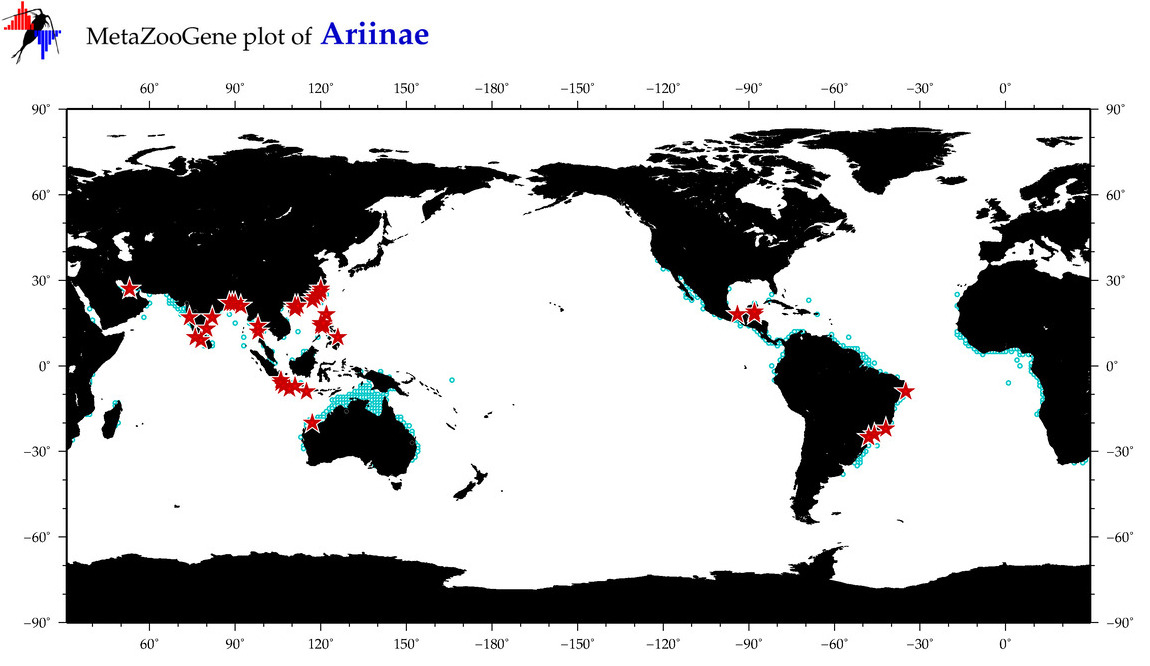
| ⋄ ⋄ Ariinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 93
Sp. w/COI-Barcodes: 37
Total COI-Barcodes: 558 |
Total Species: 93
Sp. w/COI-Barcodes: 37
Total COI-Barcodes: 558 |
 |
accepted
T5006196
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Amissidens |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014076
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Amissidens hainesi |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5016159
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Amphiarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014079
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Amphiarius rugispinis |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5016183
R:1:1:0:0 |
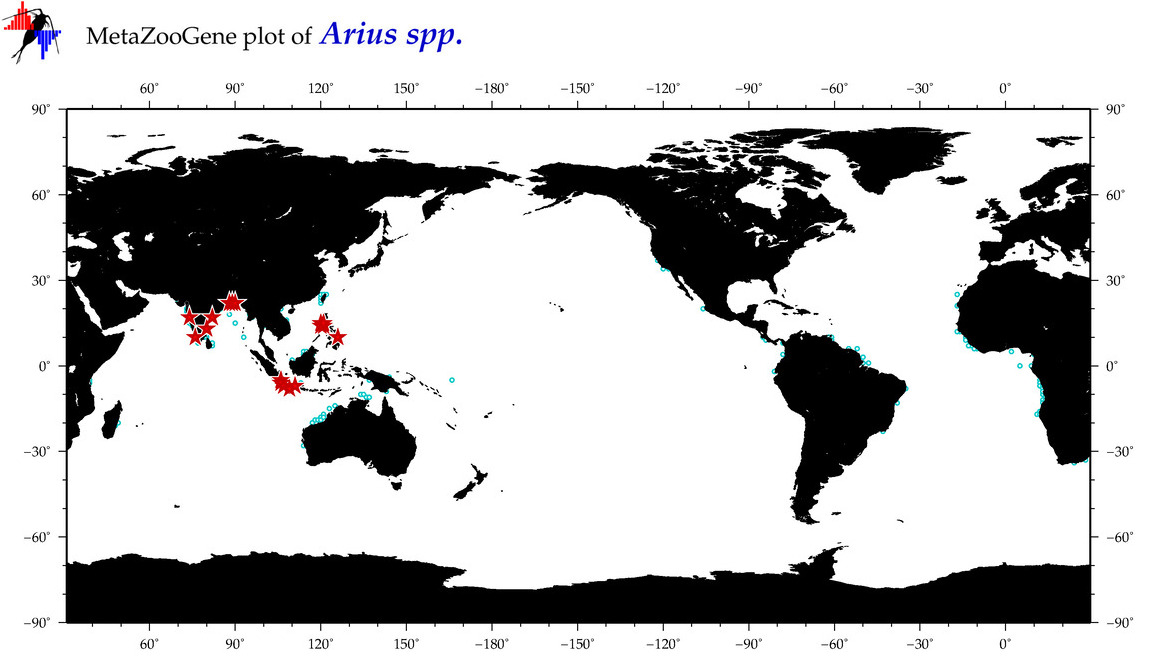
| ⋄ ⋄ ⋄ Arius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 15
Sp. w/COI-Barcodes: 10
Total COI-Barcodes: 202 |
Total Species: 15
Sp. w/COI-Barcodes: 10
Total COI-Barcodes: 202 |
 |
accepted
T5006291
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Arius arenarius |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5027713
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius arius |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 39
12S = 4
16S = 3
18S = 0
28S = 0
|
COI = 39
12S = 4
16S = 3
18S = 0
28S = 0
|
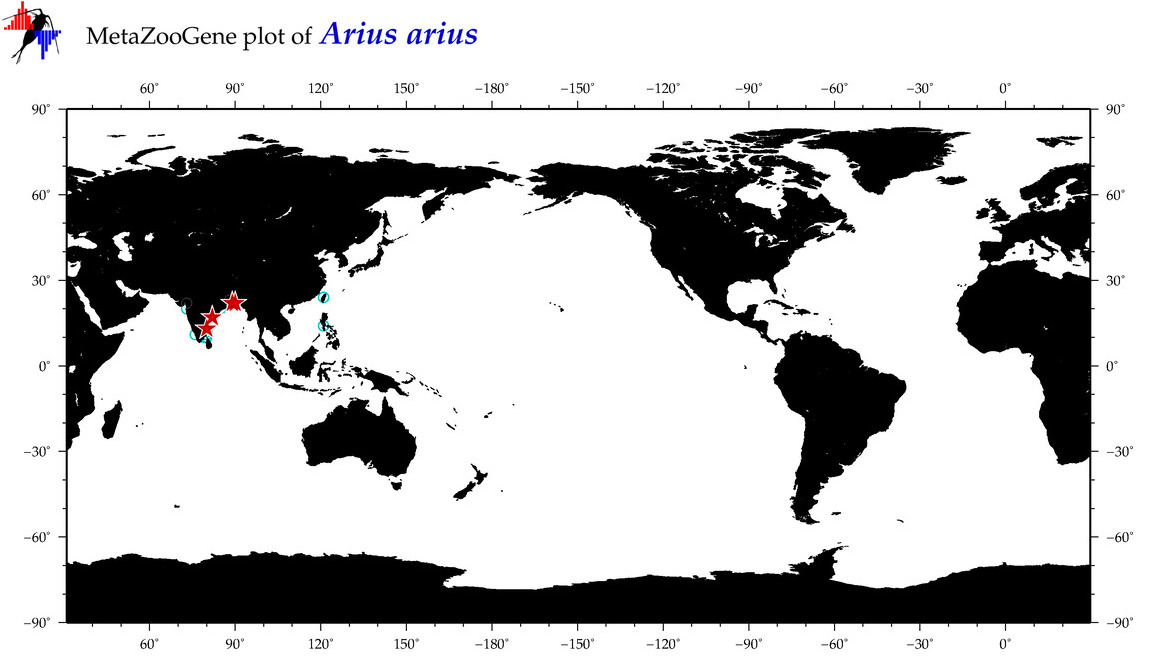 |
accepted
T5007890
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius gagora |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 11
12S = 2
16S = 1
18S = 0
28S = 0
|
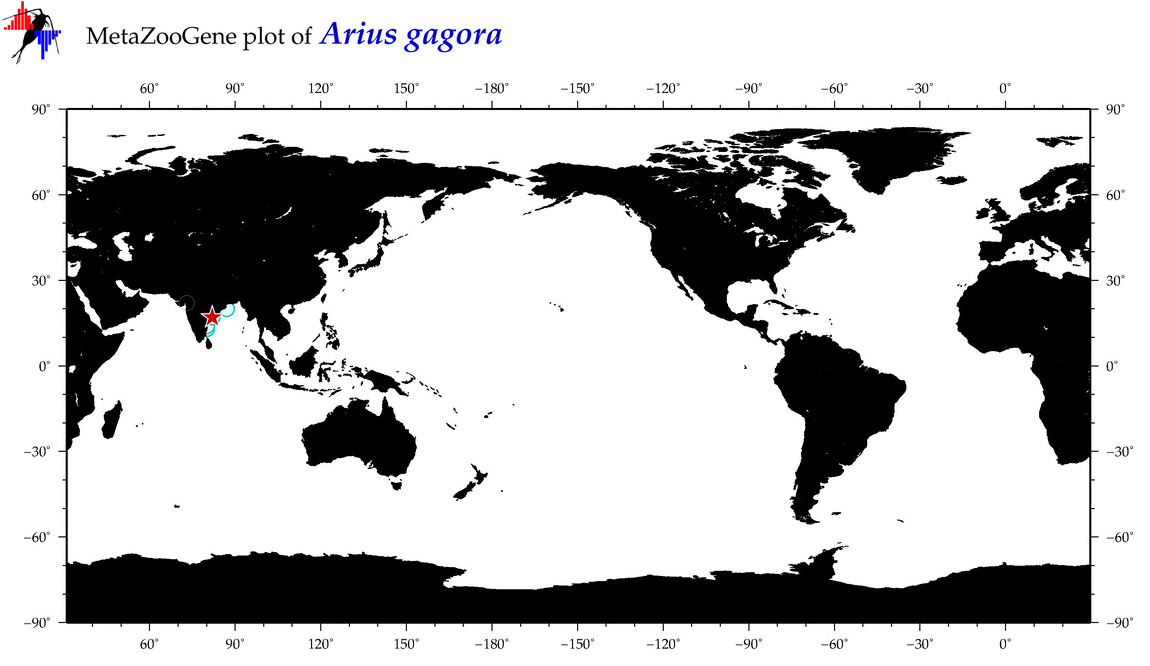 |
accepted
T5007892
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Arius jella |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
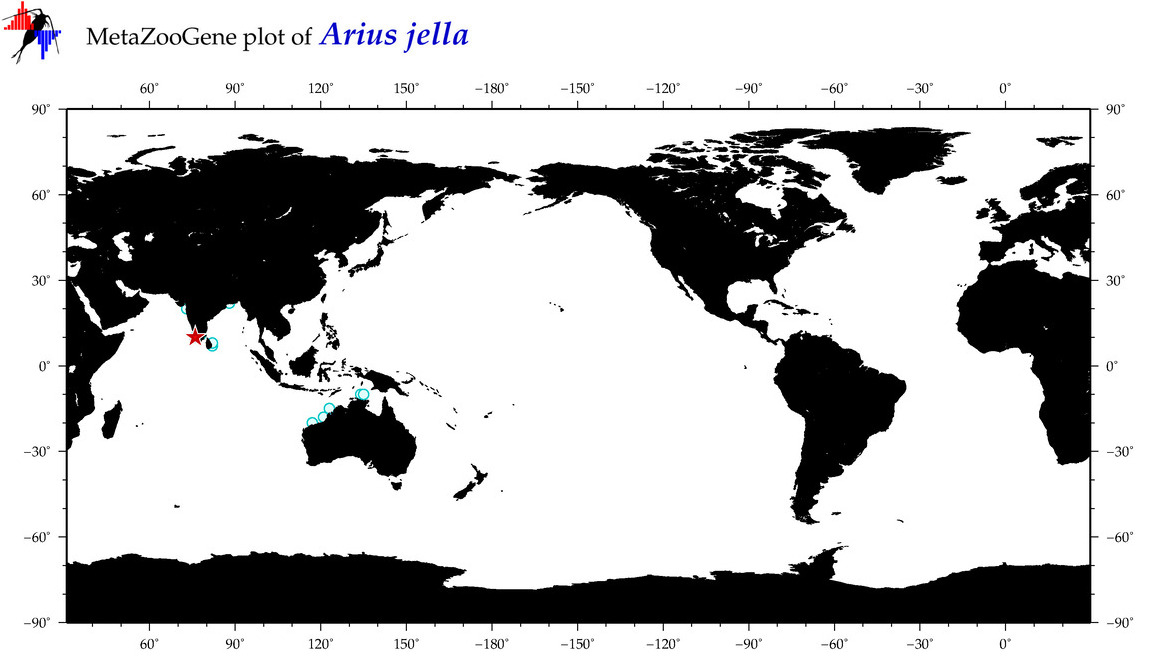 |
accepted
T5007893
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius leptonotacanthus |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 2
12S = 1
16S = 1
18S = 0
28S = 0
|
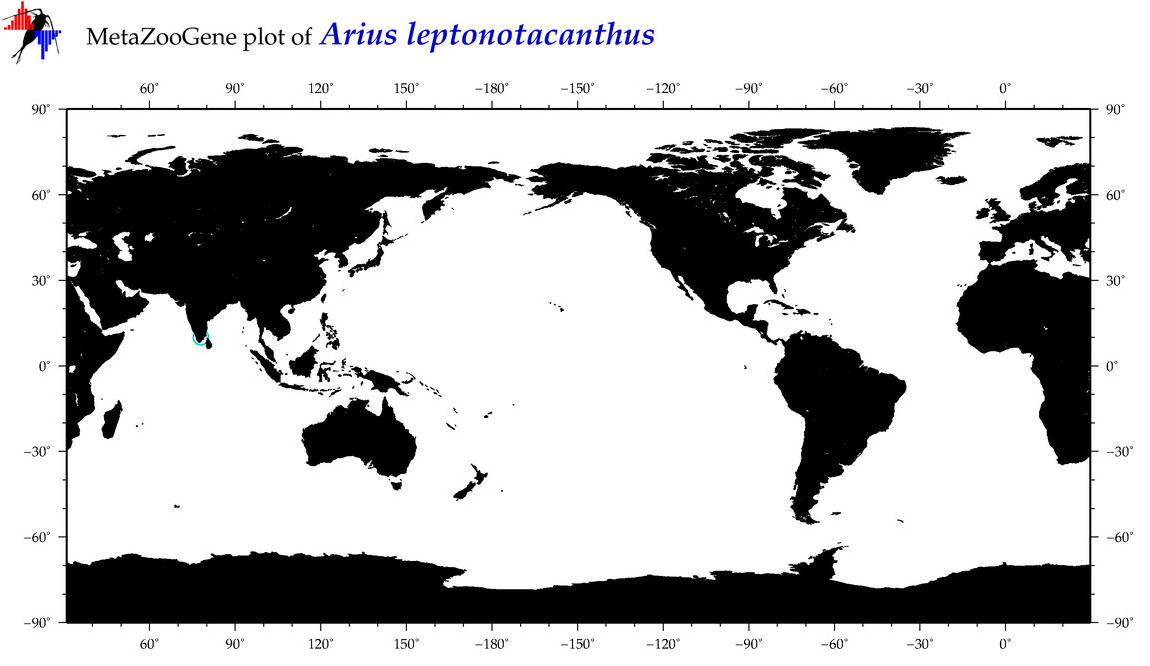 |
accepted
T5007894
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius maculatus |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 46
12S = 4
16S = 6
18S = 0
28S = 0
|
COI = 46
12S = 4
16S = 6
18S = 0
28S = 0
|
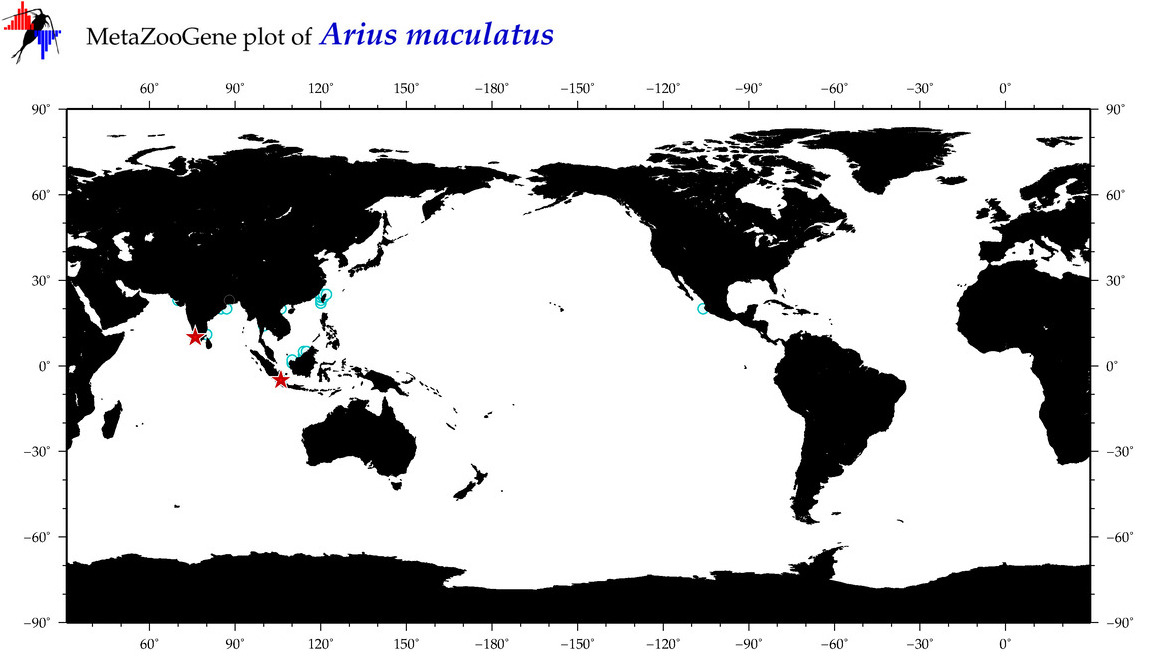 |
accepted
T5007895
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Arius madagascariensis |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5001712
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Arius manillensis |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 28
12S = 2
16S = 11
18S = 0
28S = 0
|
COI = 28
12S = 2
16S = 11
18S = 0
28S = 0
|
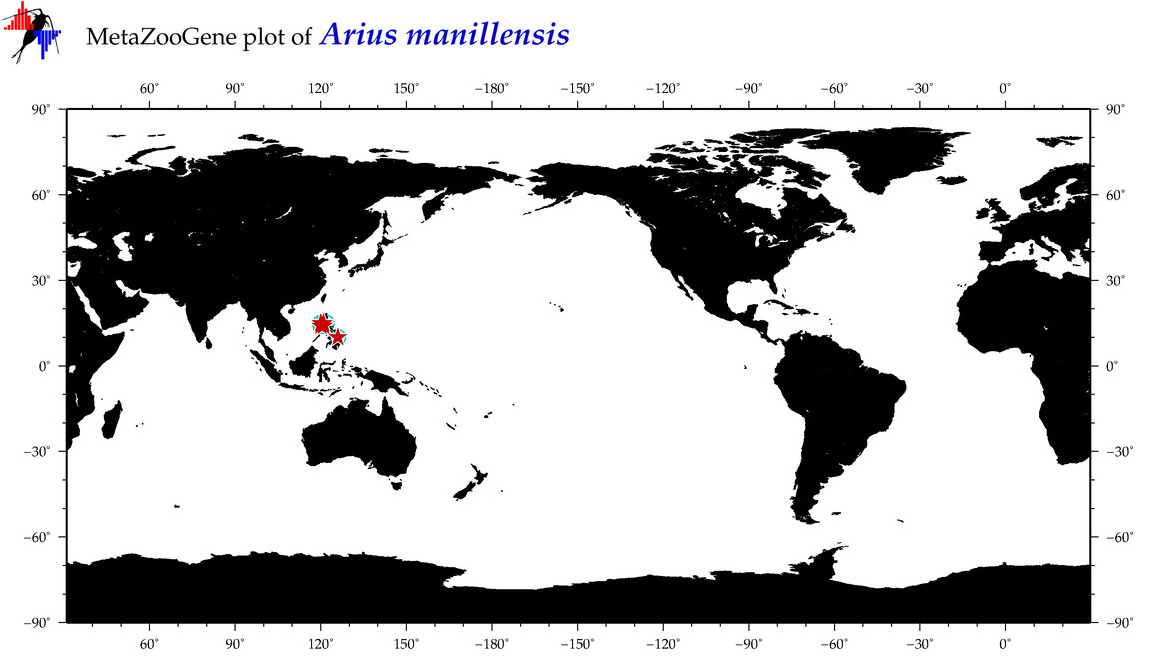 |
accepted
T5007896
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Arius microcephalus |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016636
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius oetik |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 2
16S = 1
18S = 0
28S = 0
|
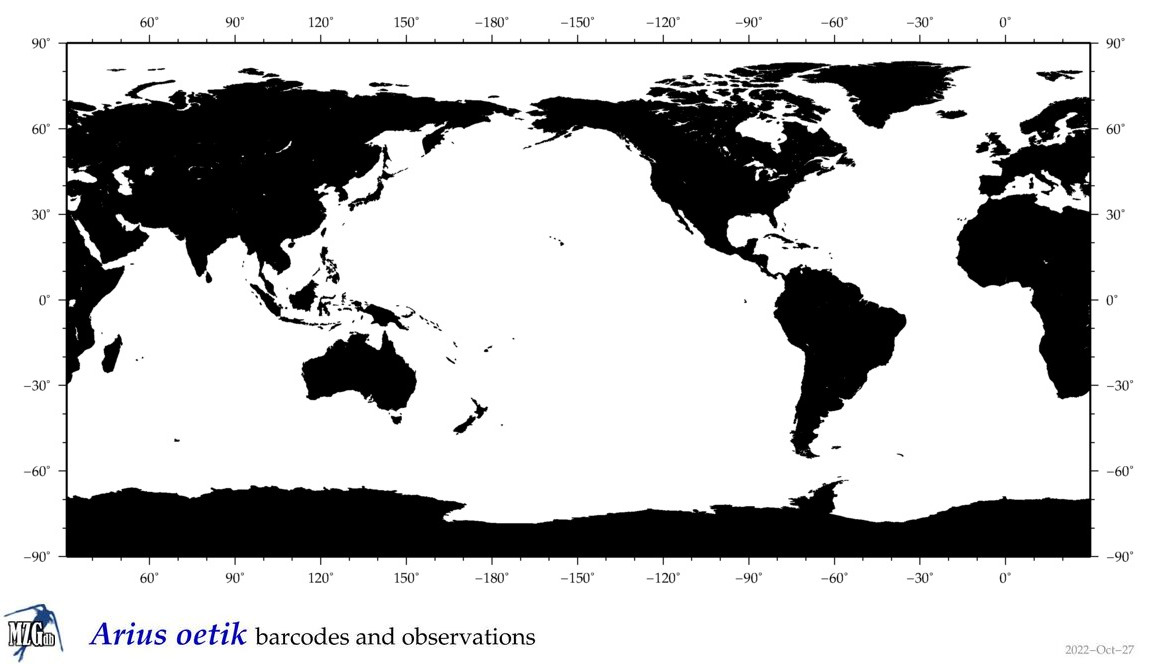 |
accepted
T5027714
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius subrostratus |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 35
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 35
12S = 0
16S = 2
18S = 0
28S = 0
|
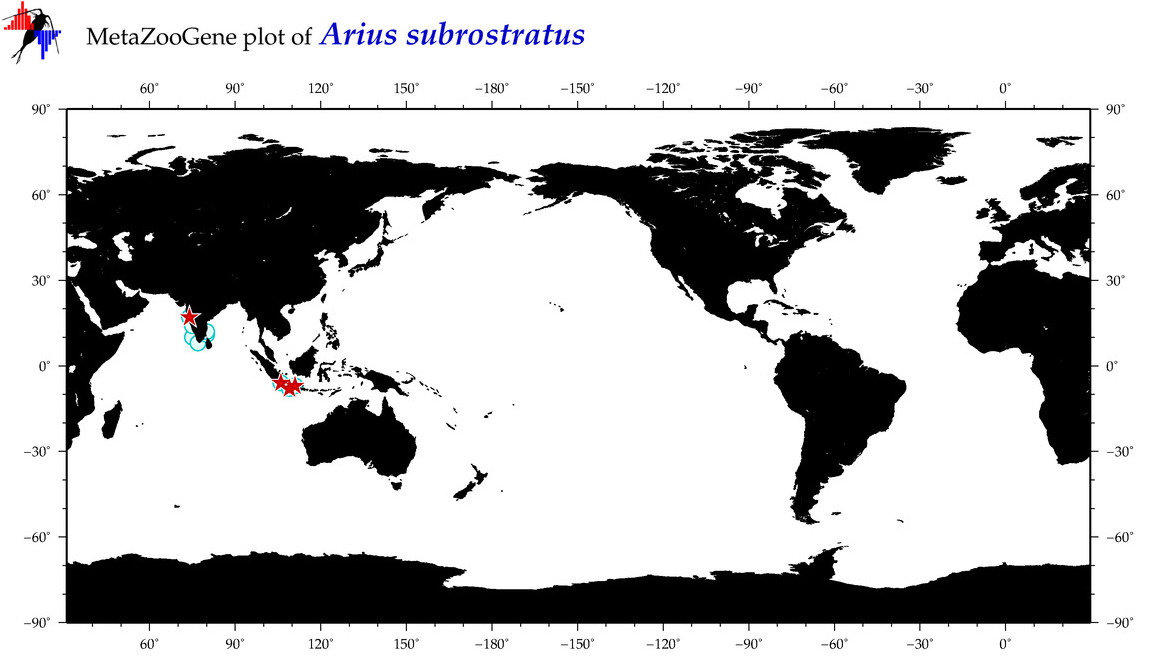 |
accepted
T5007897
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius sumatranus |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016639
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Arius venosus |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 6
12S = 1
16S = 0
18S = 0
28S = 0
|
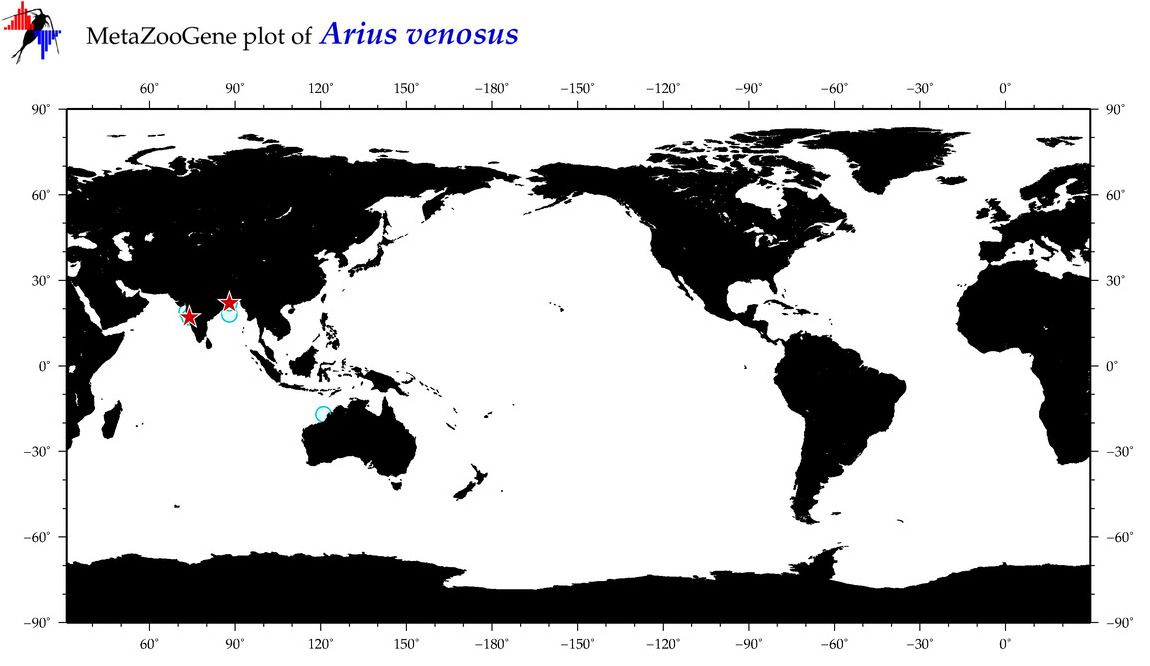 |
accepted
T5007898
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Aspistor |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 7 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 7 |
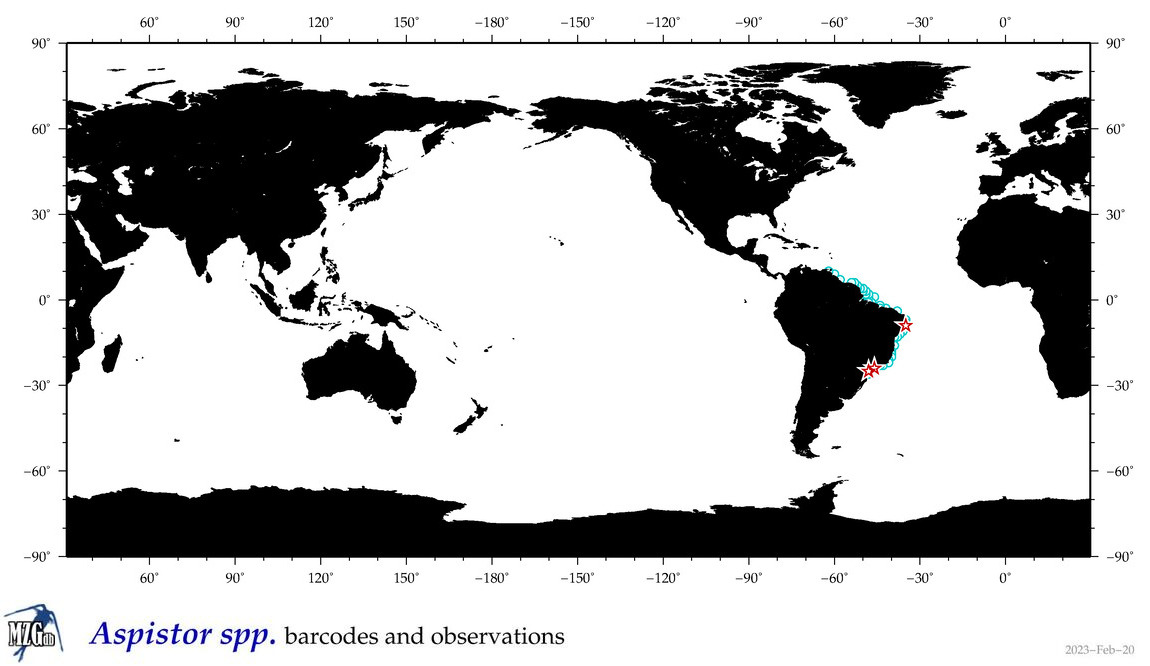 |
accepted
T5014129
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Aspistor luniscutis |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 7
12S = 2
16S = 2
18S = 0
28S = 0
|
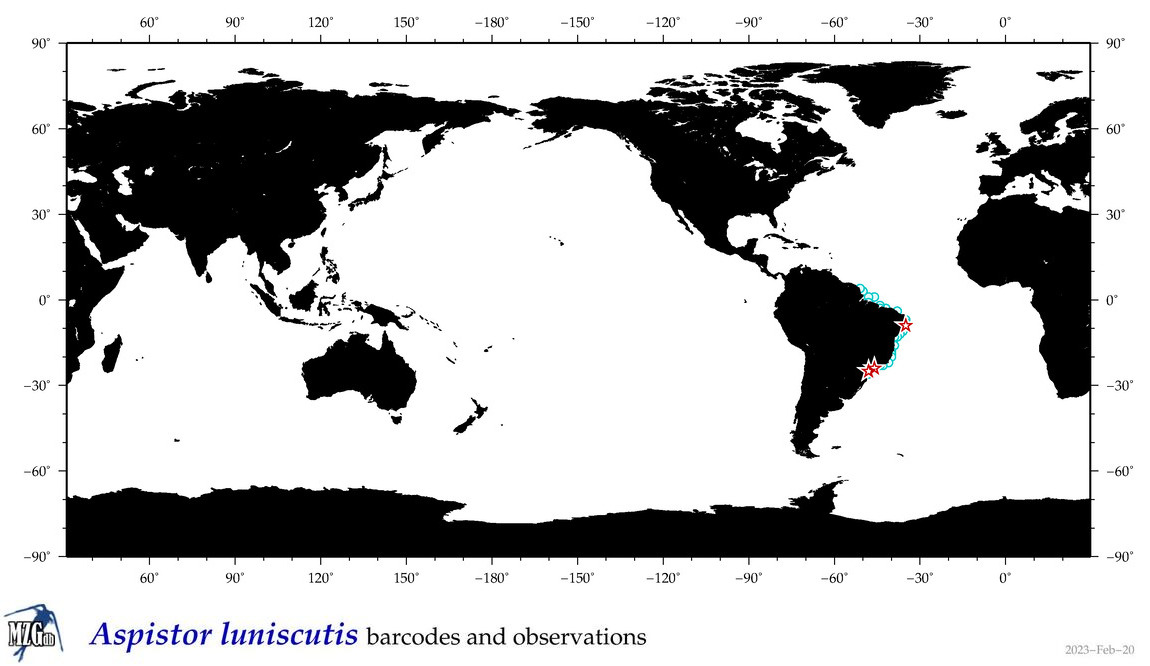 |
accepted
T5016719
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Aspistor quadriscutis |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5016720
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Carlarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 3
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014237
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Carlarius heudelotii |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017646
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Carlarius latiscutatus |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
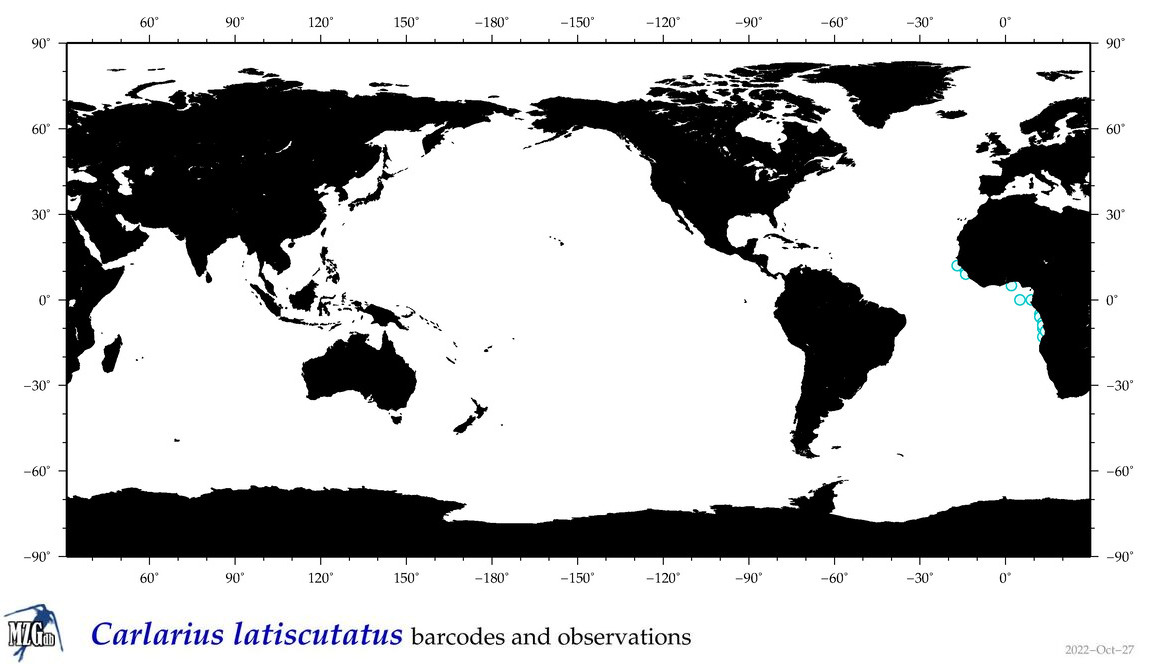 |
accepted
T5027827
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Carlarius parkii |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
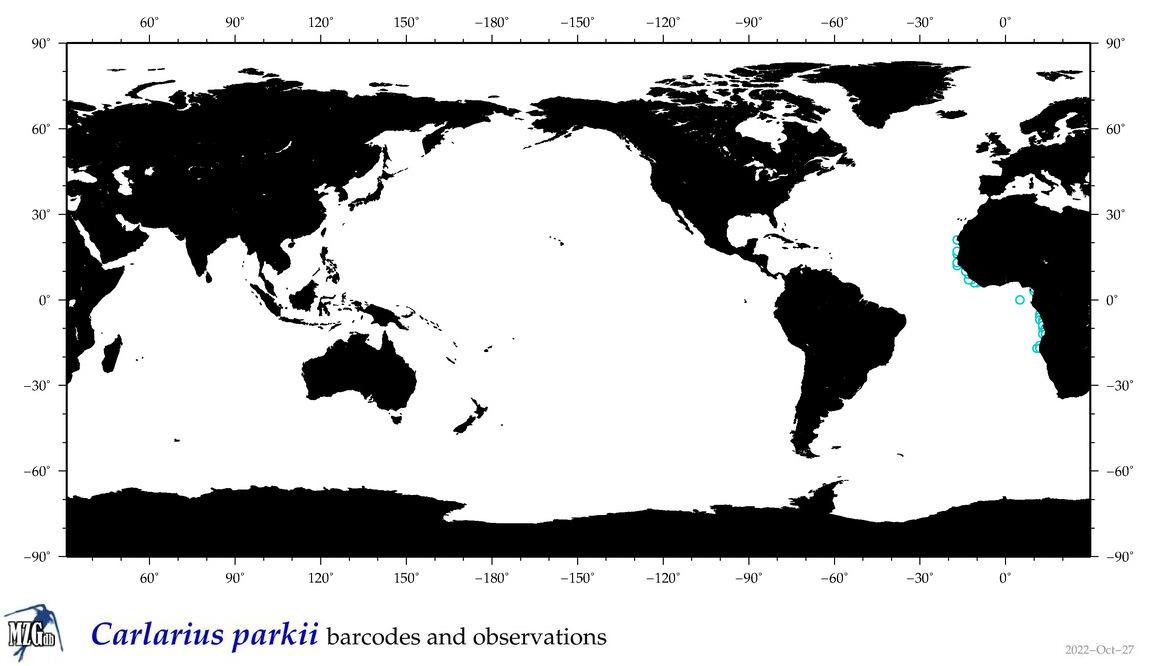 |
accepted
T5027828
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Cathorops |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 11
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 16 |
Total Species: 11
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 16 |
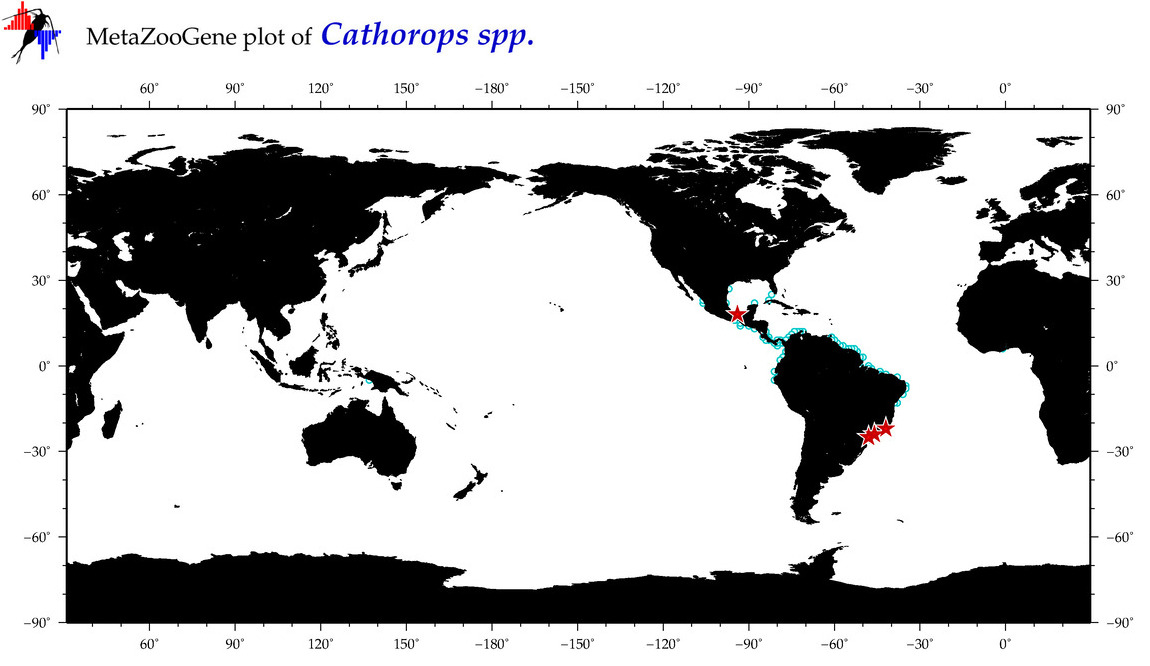 |
accepted
T5006579
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops dasycephalus |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017659
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops fuerthii |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017660
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops hypophthalmus |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017661
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops multiradiatus |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017665
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops spixii |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 16
12S = 1
16S = 2
18S = 0
28S = 0
|
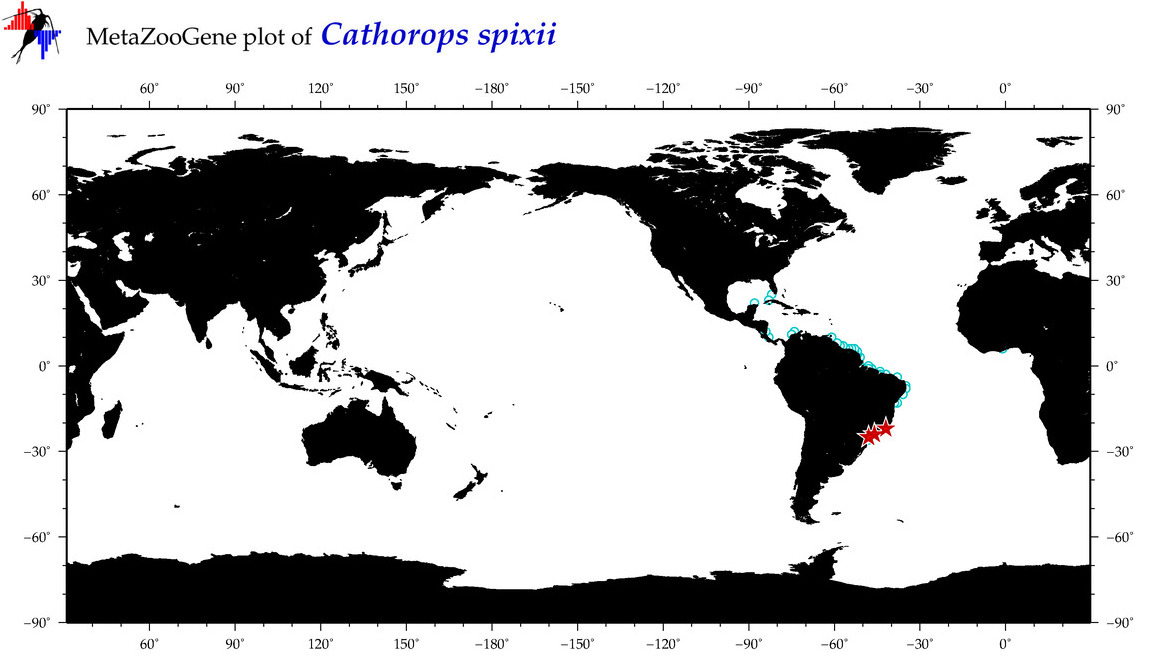 |
accepted
T5008368
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops steindachneri |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017666
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops tuyra |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5017667
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cathorops wayuu |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5013490
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Cephalocassis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
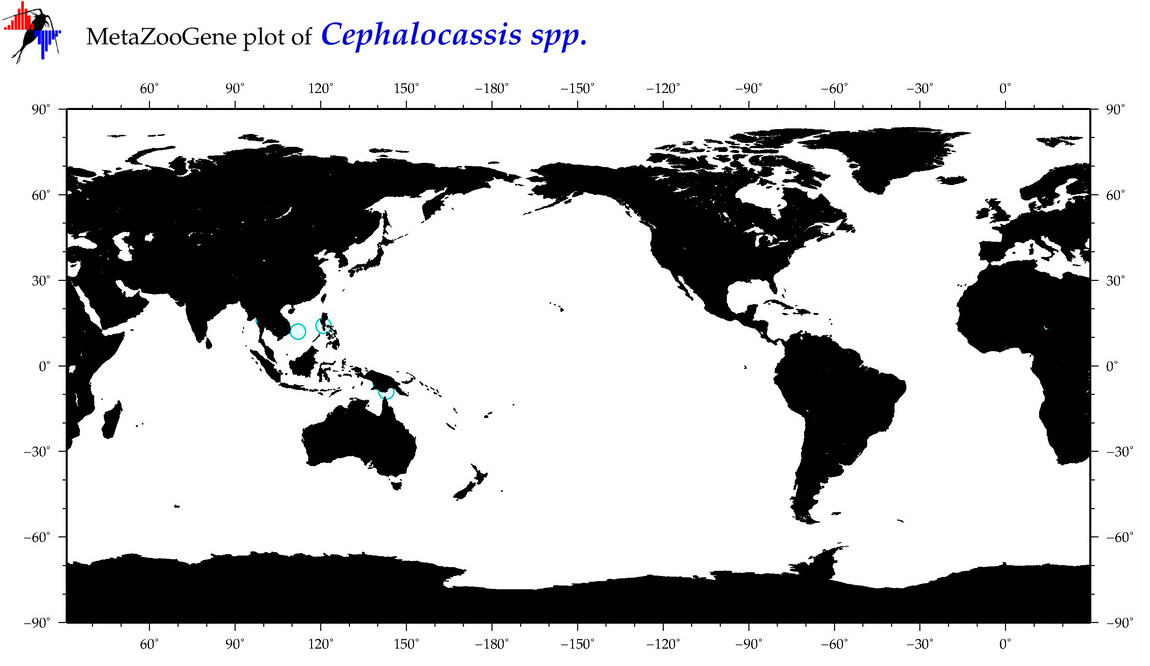 |
accepted
T5006584
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cephalocassis jatia |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
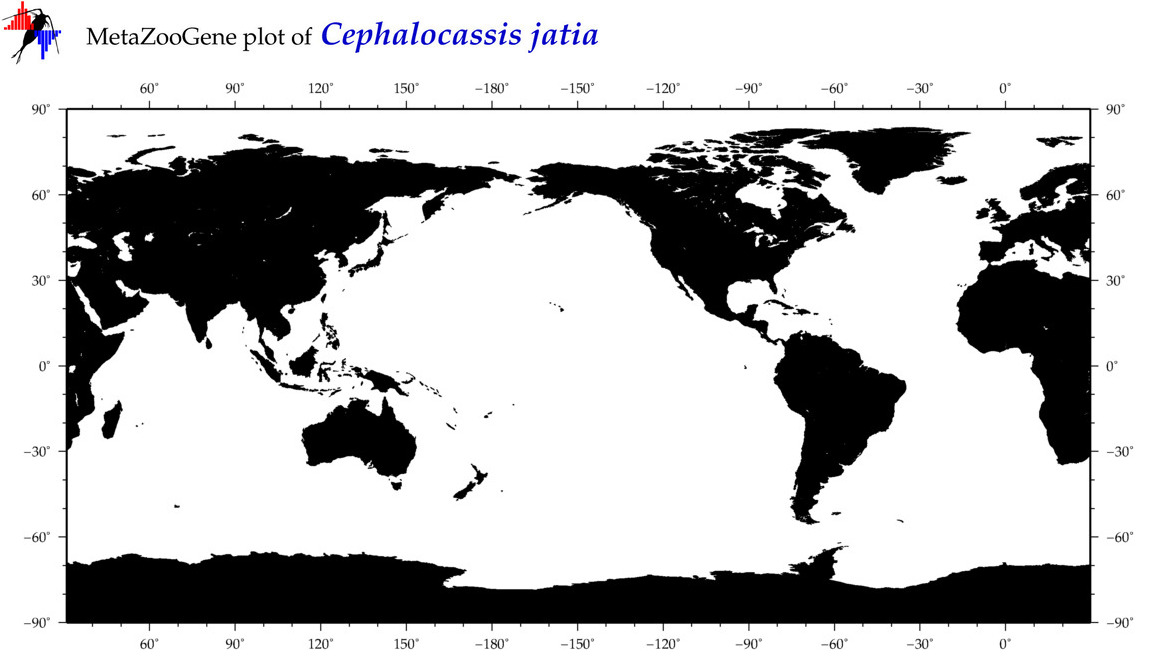 |
accepted
T5008388
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Cephalocassis melanochir |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5013504
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Cinetodus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014286
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Cochlefelis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014305
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Cryptarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
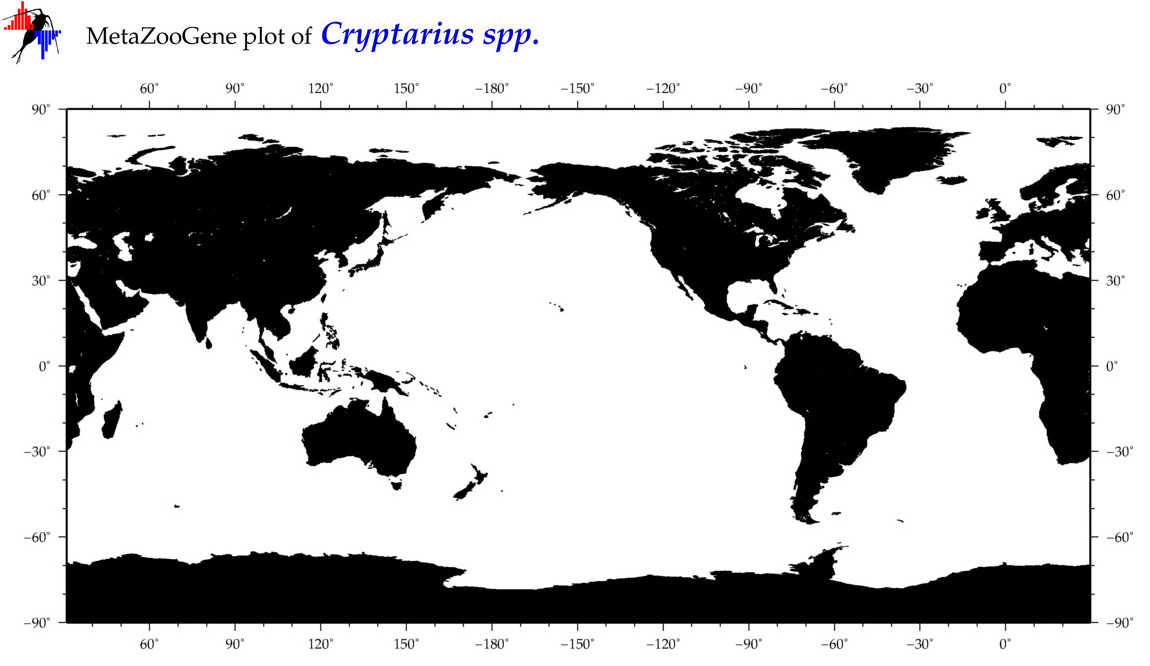 |
accepted
T5006646
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Genidens |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 21 |
Total Species: 4
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 21 |
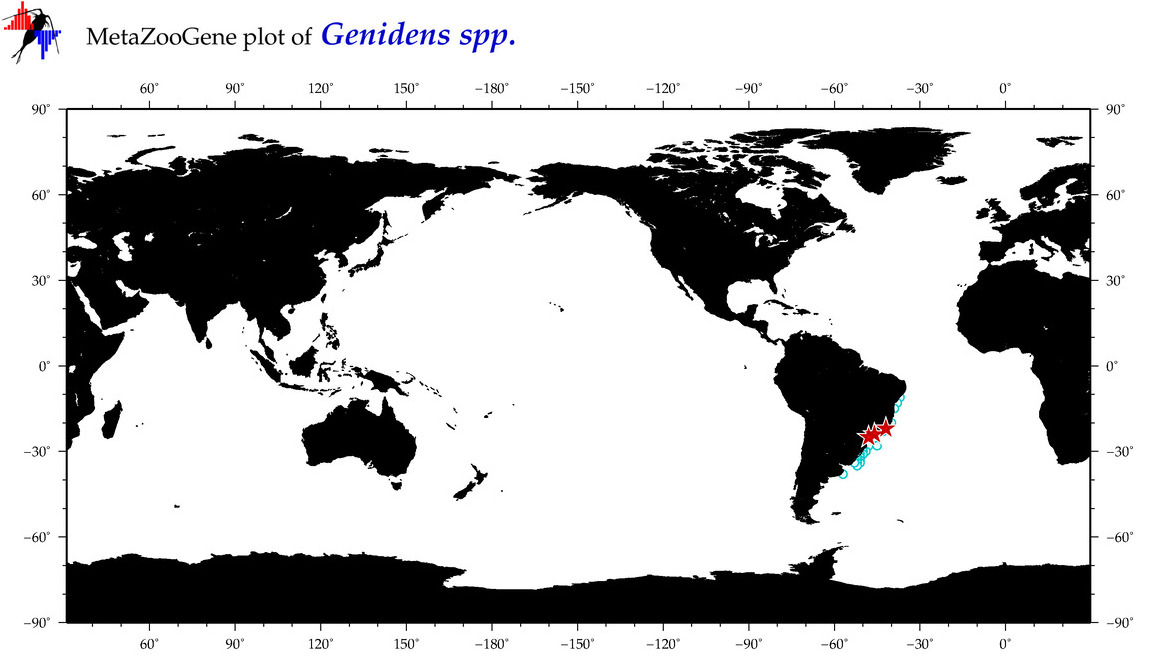 |
accepted
T5006736
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Genidens barbus |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 15
12S = 1
16S = 2
18S = 0
28S = 0
|
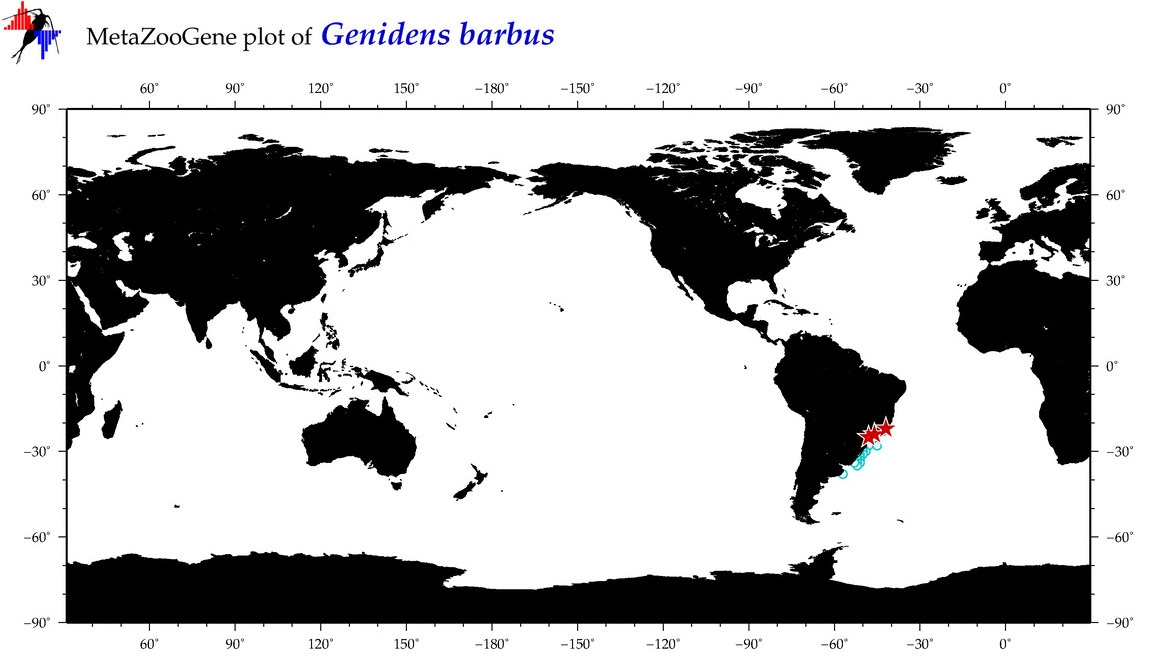 |
accepted
T5009336
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Genidens genidens |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 6
12S = 1
16S = 2
18S = 0
28S = 0
|
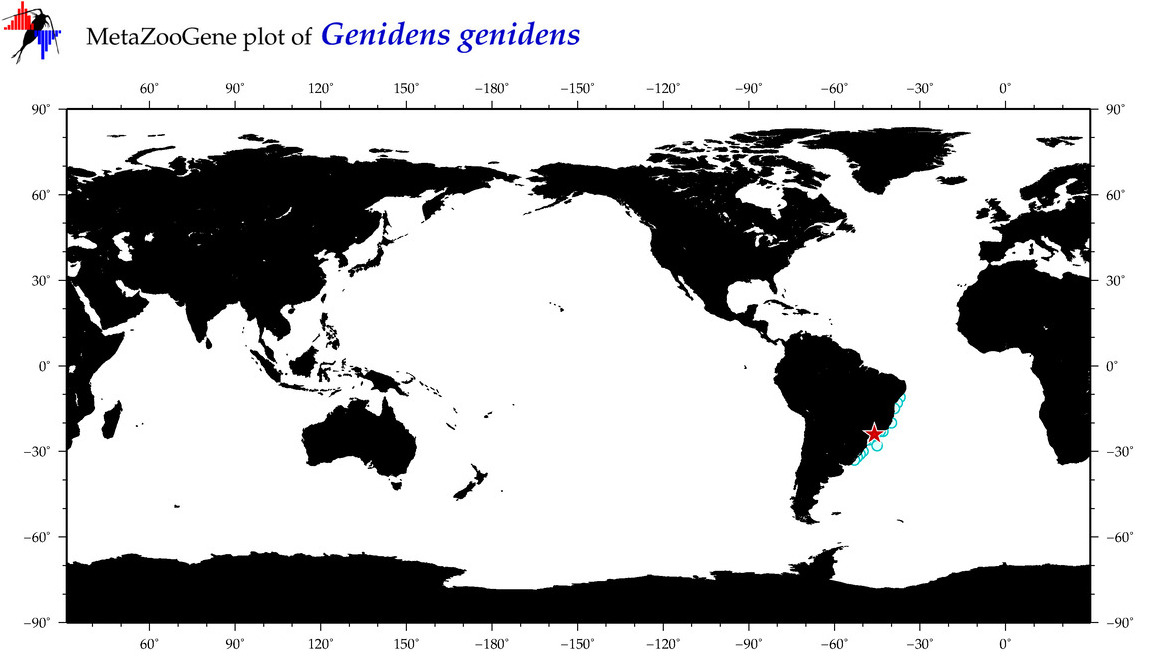 |
accepted
T5009337
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Genidens machadoi |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5027398
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Genidens planifrons |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5020057
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Hemiarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
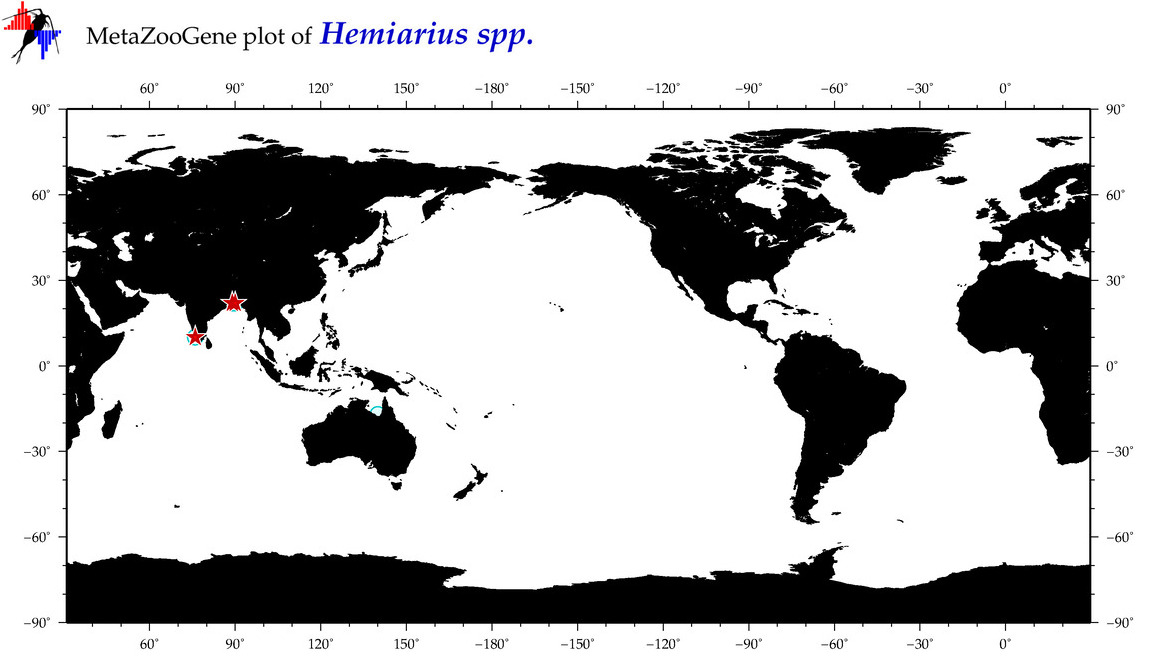 |
accepted
T5006777
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Hemiarius dioctes |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 3
18S = 0
28S = 0
|
no map |
accepted
T5020680
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Hemiarius stormii |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
 |
accepted
T5028088
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Hexanematichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 13 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 13 |
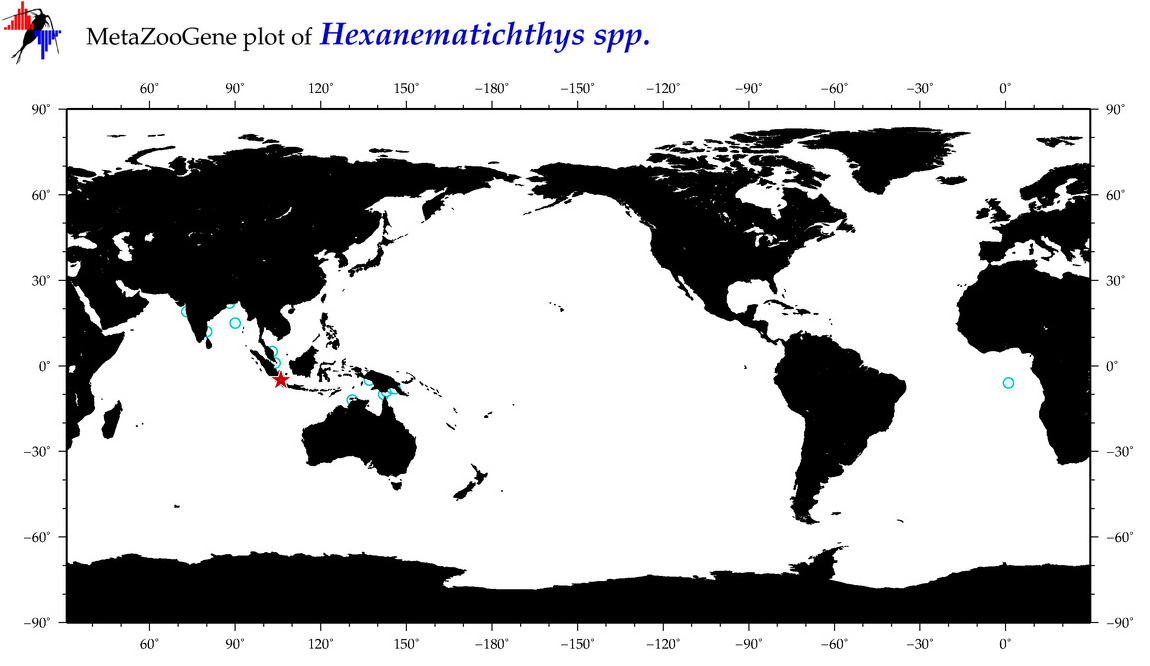 |
accepted
T5006805
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Hexanematichthys mastersi |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5020820
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hexanematichthys sagor |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 13
12S = 2
16S = 1
18S = 0
28S = 0
|
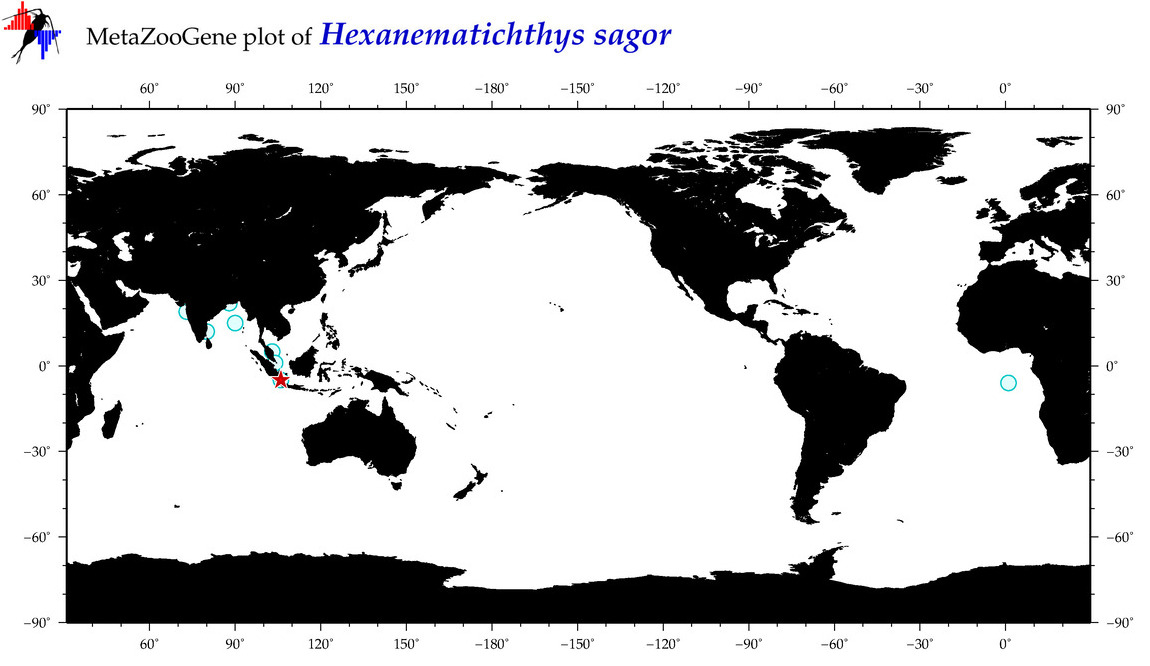 |
accepted
T5009652
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Ketengus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014617
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Ketengus typus |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5021250
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Nedystoma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
no map |
accepted
T5014777
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Nemapteryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 21 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 21 |
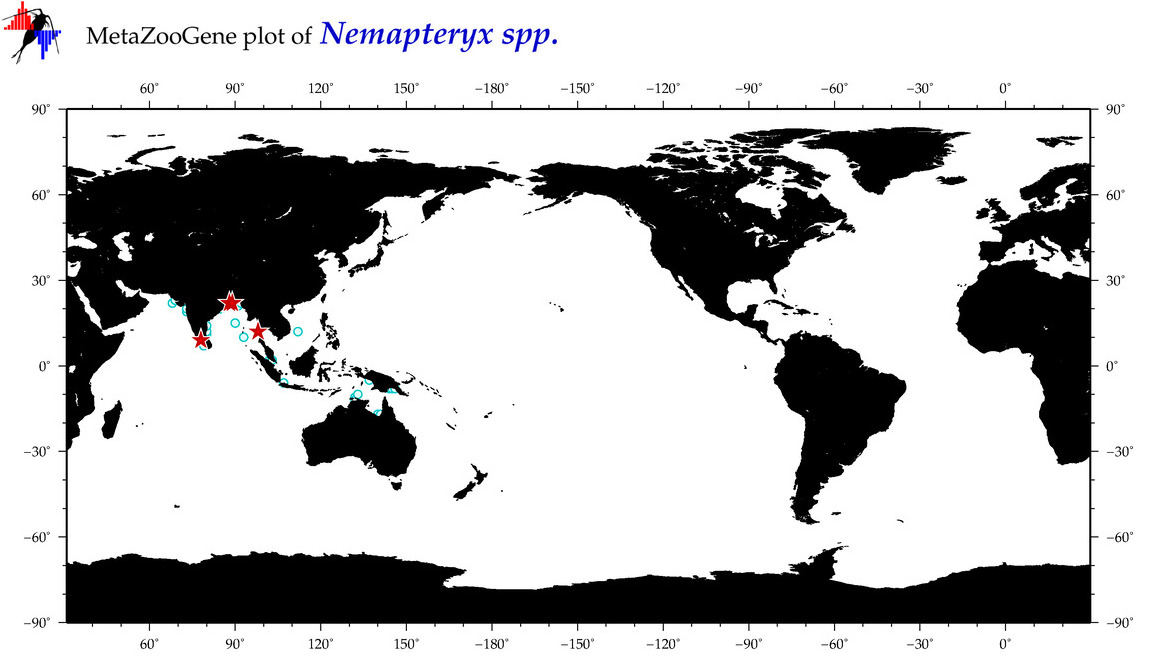 |
accepted
T5006990
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Nemapteryx armiger |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022510
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Nemapteryx caelata |
Species
(41) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 15
12S = 1
16S = 0
18S = 0
28S = 0
|
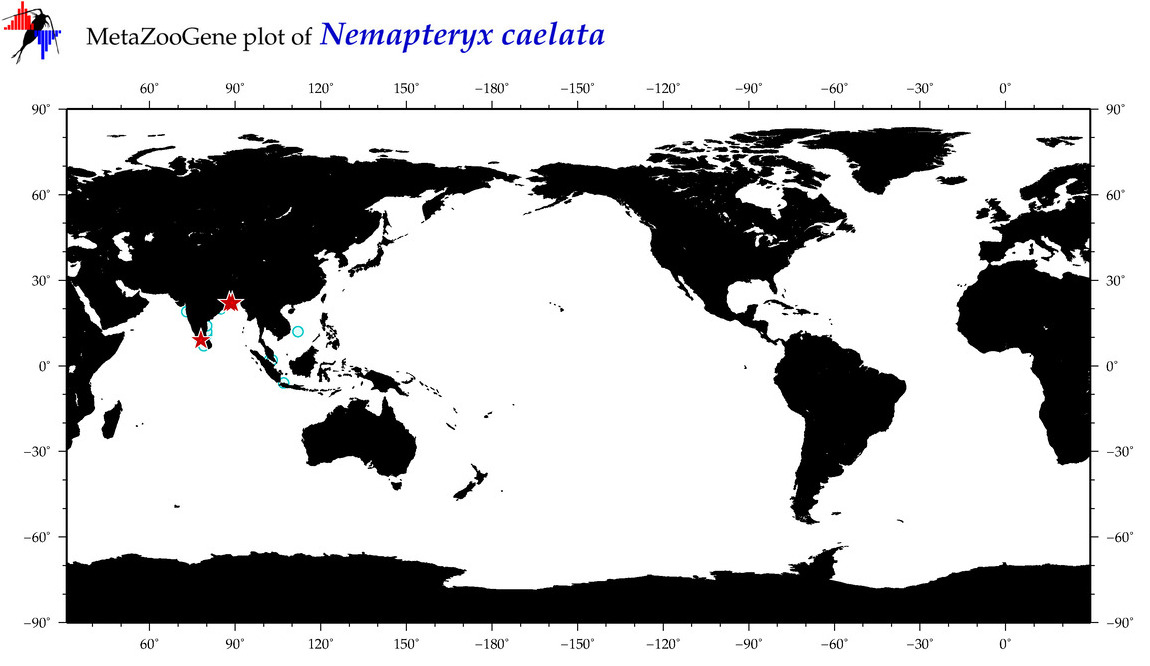 |
accepted
T5010604
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Nemapteryx macronotacantha |
Species
(42) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022511
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Nemapteryx nenga |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 5
12S = 2
16S = 1
18S = 0
28S = 0
|
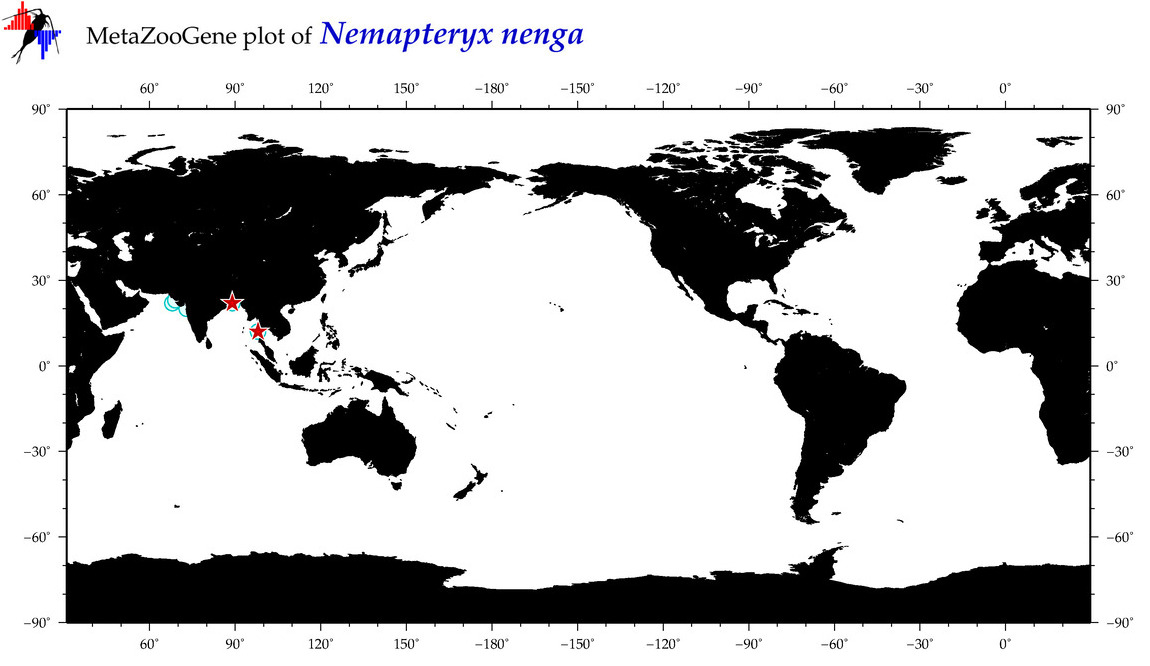 |
accepted
T5010605
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Neoarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 13 |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 13 |
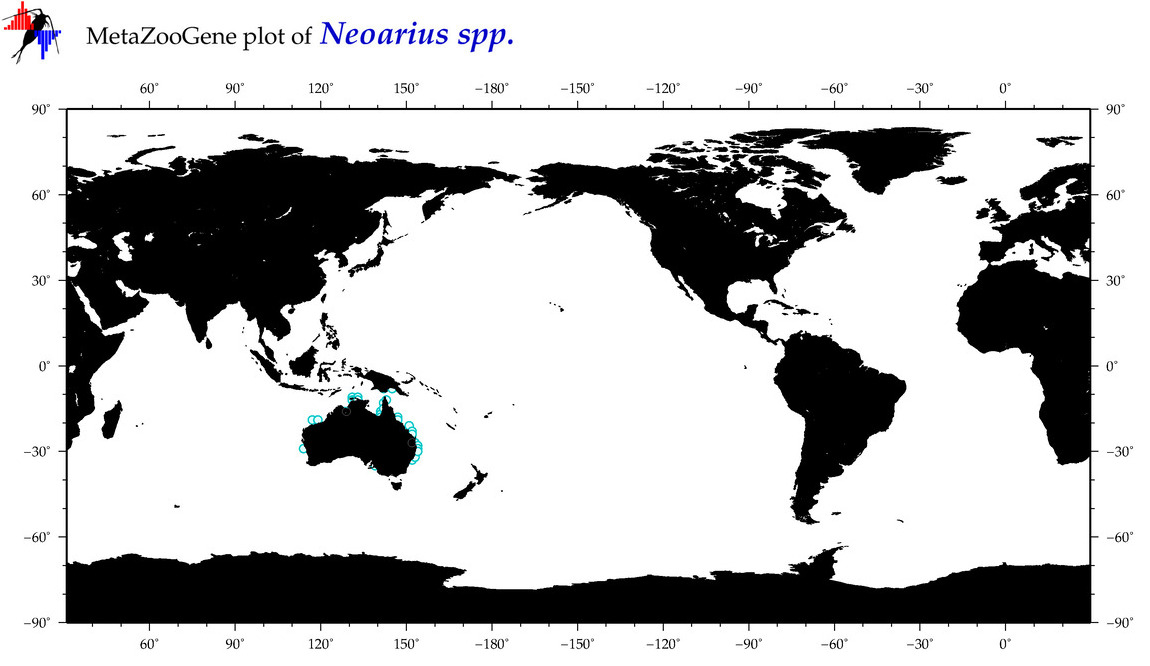 |
accepted
T5006996
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Neoarius graeffei |
Species
(44) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 2
16S = 1
18S = 1
28S = 0
|
COI = 13
12S = 2
16S = 1
18S = 1
28S = 0
|
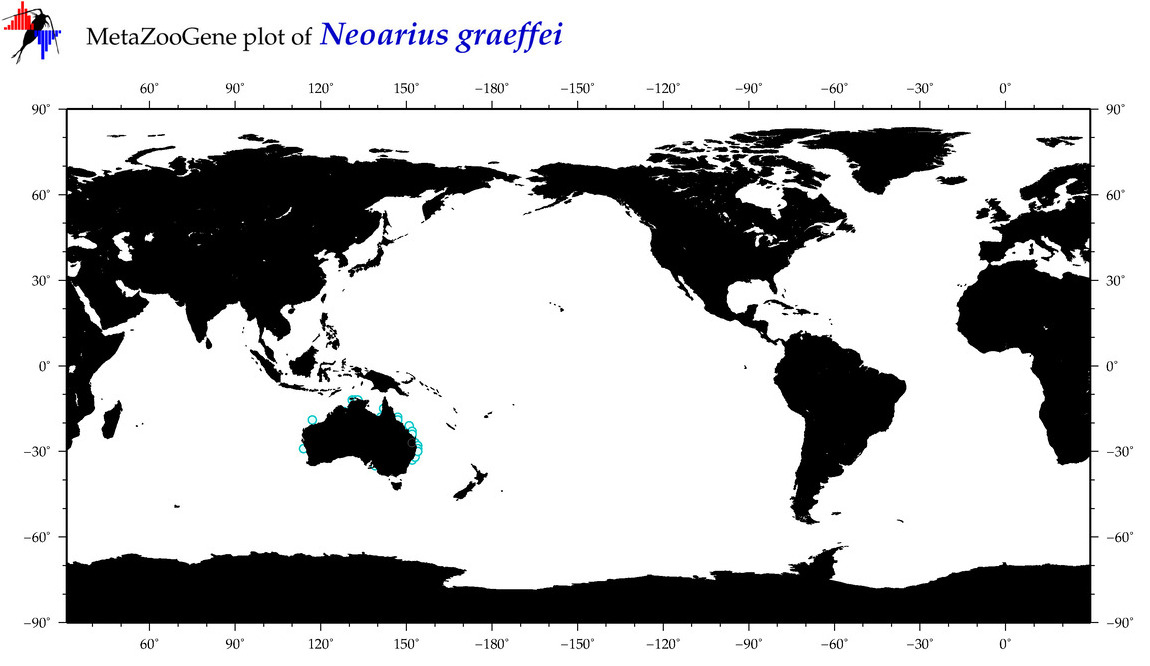 |
accepted
T5010613
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Neoarius leptaspis |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022550
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Netuma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 119 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 119 |
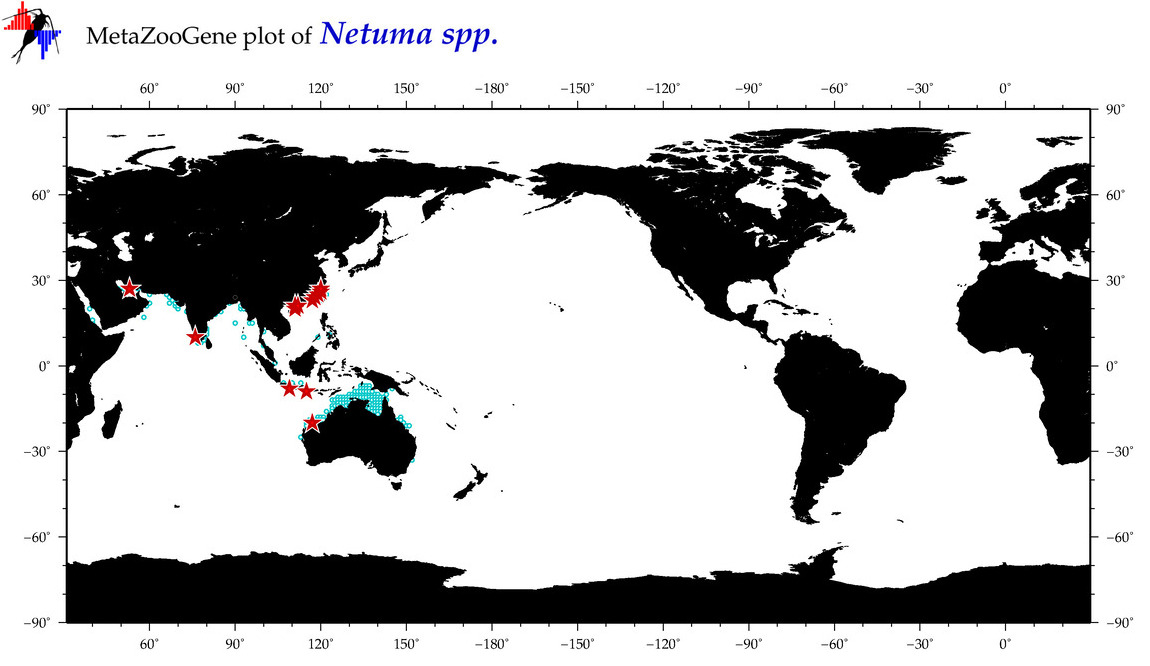 |
accepted
T5007010
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Netuma bilineata |
Species
(46) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 9
12S = 2
16S = 1
18S = 0
28S = 0
|
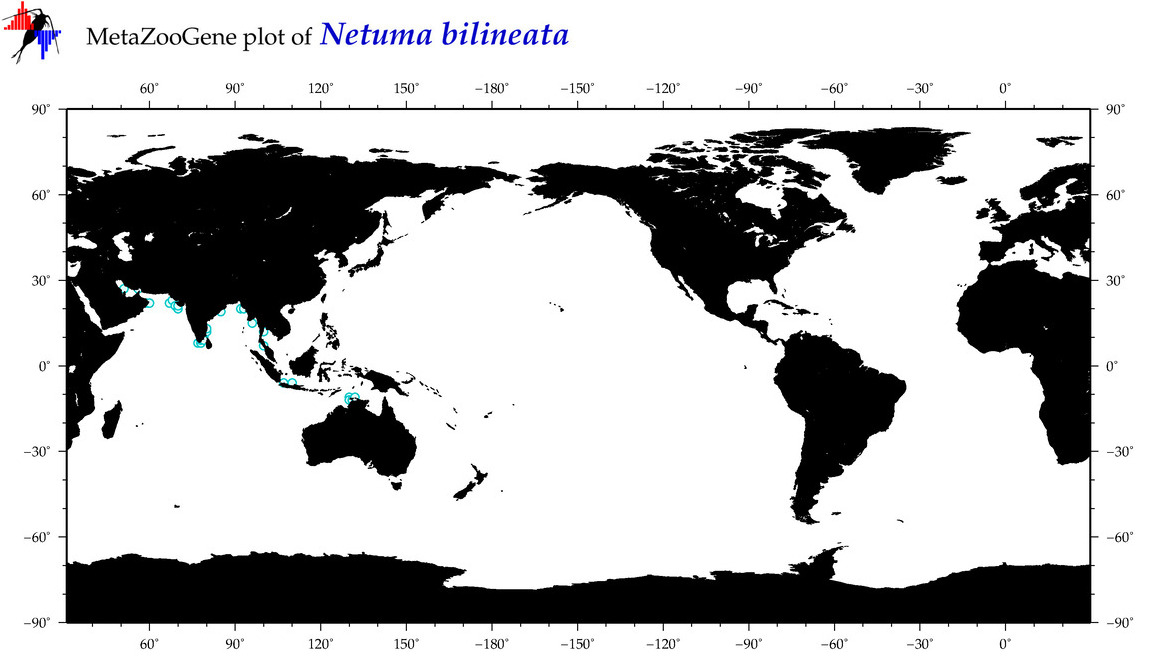 |
accepted
T5010685
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Netuma patriciae |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
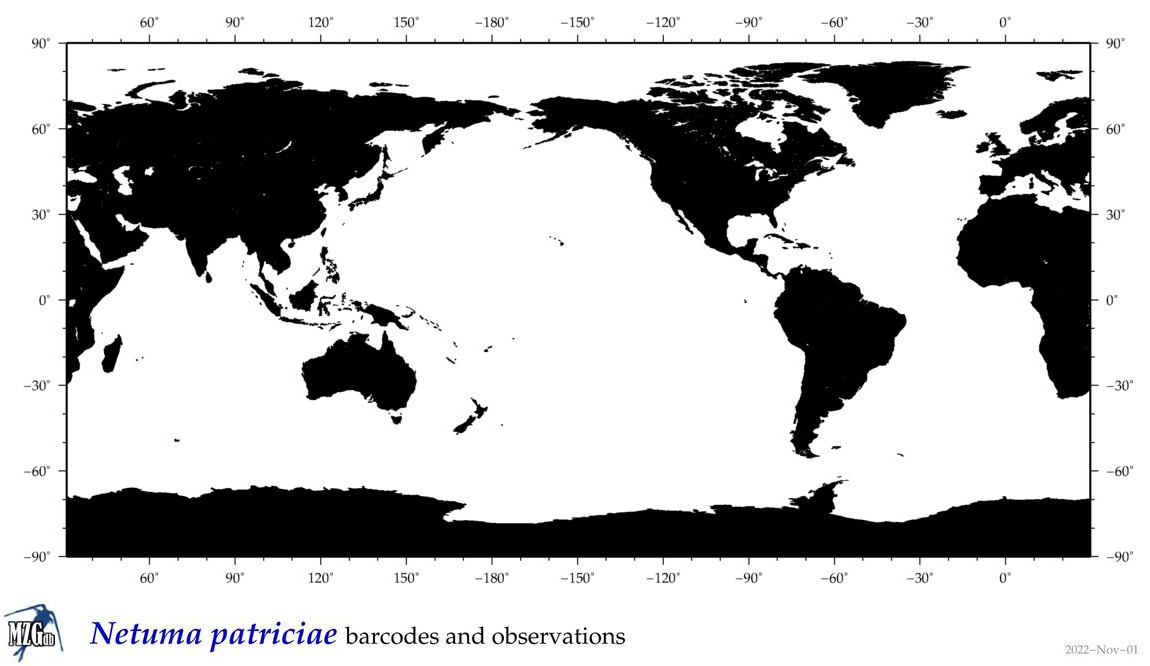 |
accepted
T5032278
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Netuma proxima |
Species
(48) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022694
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Netuma thalassina |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 109
12S = 7
16S = 11
18S = 0
28S = 0
|
COI = 109
12S = 7
16S = 11
18S = 0
28S = 0
|
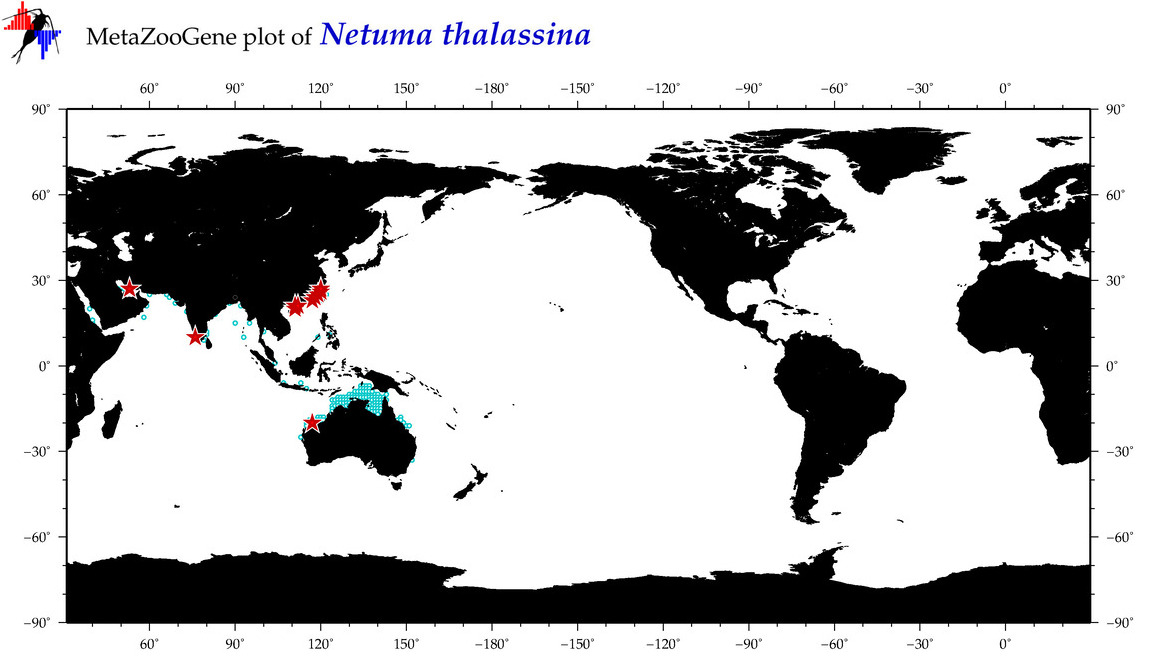 |
accepted
T5010686
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Notarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 12
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 6 |
Total Species: 12
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 6 |
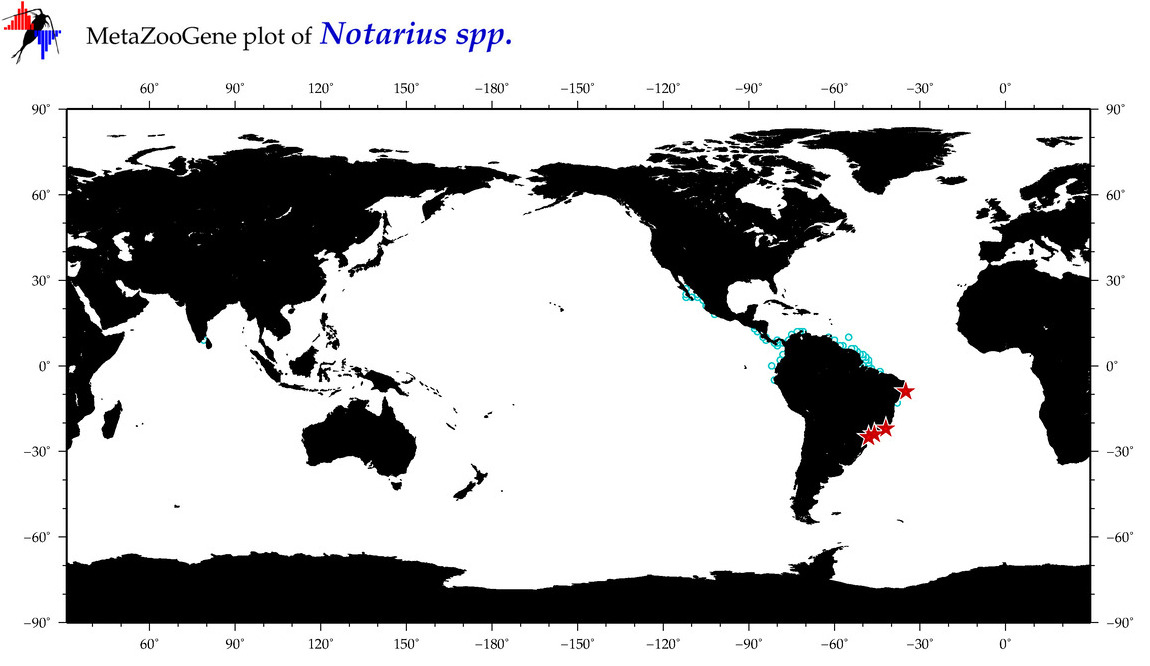 |
accepted
T5007014
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius armbrusteri |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022756
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius biffi |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022757
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius grandicassis |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 6
12S = 1
16S = 1
18S = 0
28S = 0
|
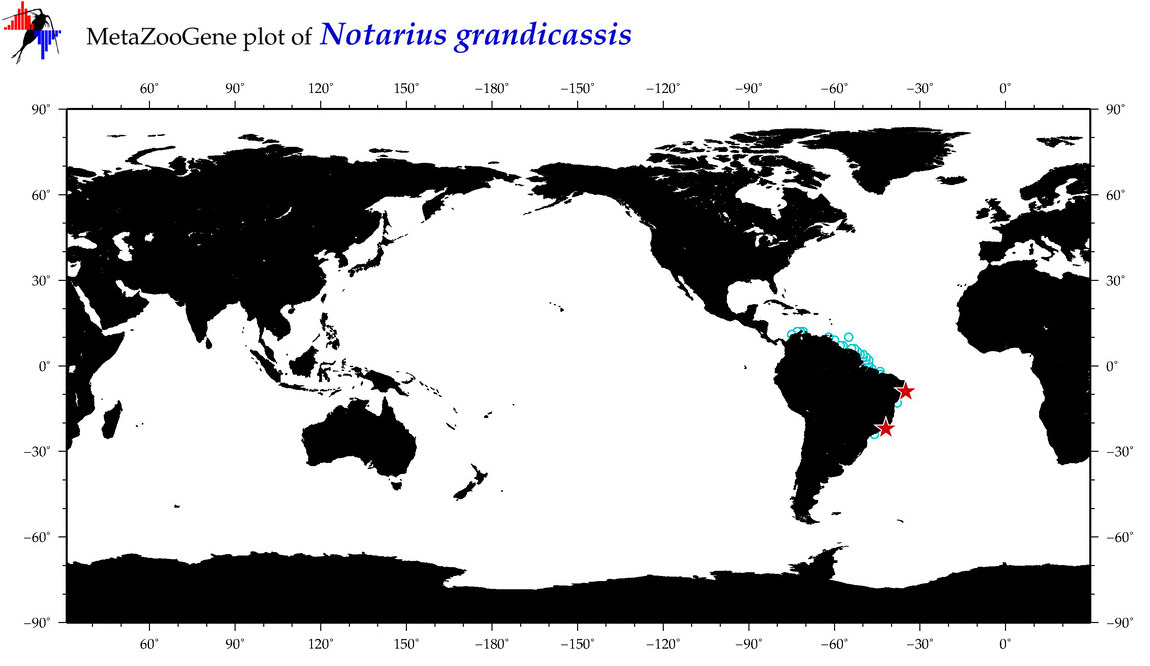 |
accepted
T5010699
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius insculptus |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022759
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius kessleri |
Species
(54) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022760
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius lentiginosus |
Species
(55) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022761
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius neogranatensis |
Species
(56) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022762
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius osculus |
Species
(57) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022763
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius planiceps |
Species
(58) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022764
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notarius troschelii |
Species
(59) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5022765
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Occidentarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
no map |
accepted
T5014816
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Occidentarius platypogon |
Species
(60) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 3
16S = 3
18S = 0
28S = 0
|
COI = 2
12S = 3
16S = 3
18S = 0
28S = 0
|
no map |
accepted
T5022830
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Osteogeneiosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 28 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 28 |
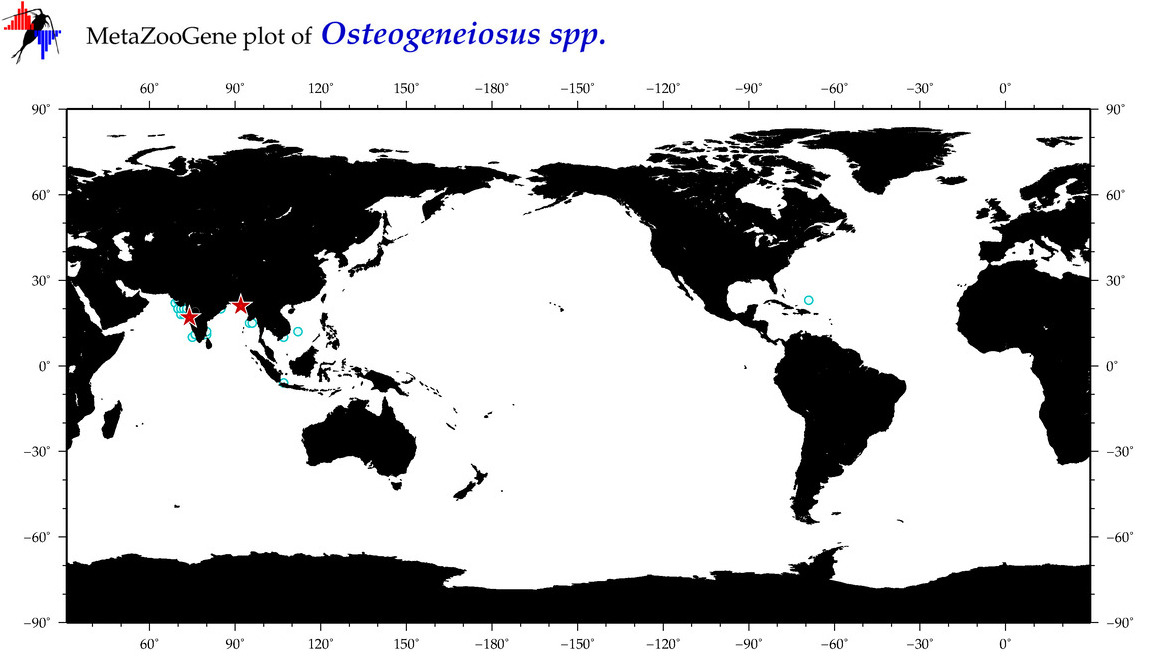 |
accepted
T5007050
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Osteogeneiosus militaris |
Species
(61) |
COI
12S
16S
18S
28S
|
COI = 28
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 28
12S = 2
16S = 2
18S = 0
28S = 0
|
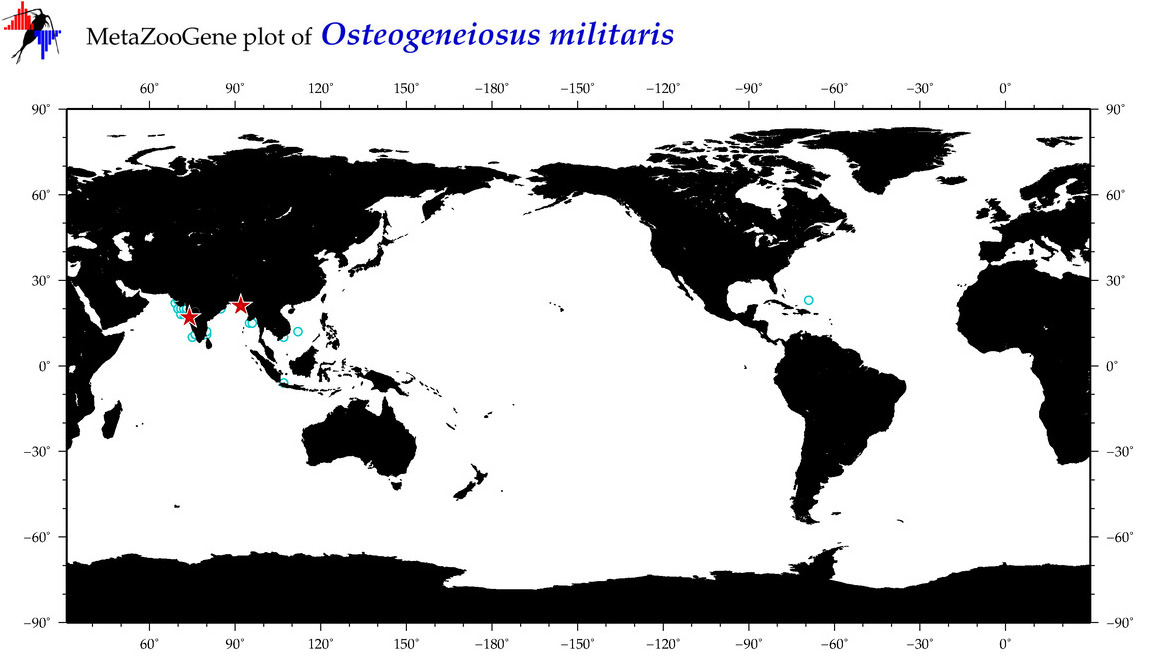 |
accepted
T5011001
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Plicofollis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 8
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 65 |
Total Species: 8
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 65 |
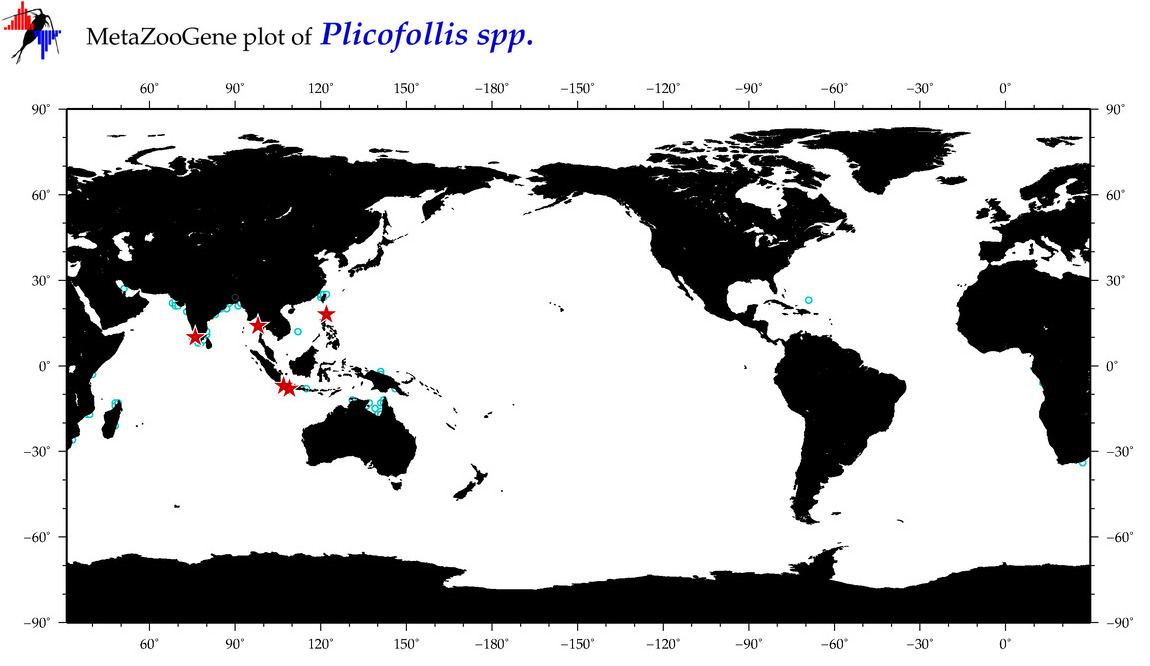 |
accepted
T5007161
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis argyropleuron |
Species
(62) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 18
12S = 3
16S = 2
18S = 0
28S = 0
|
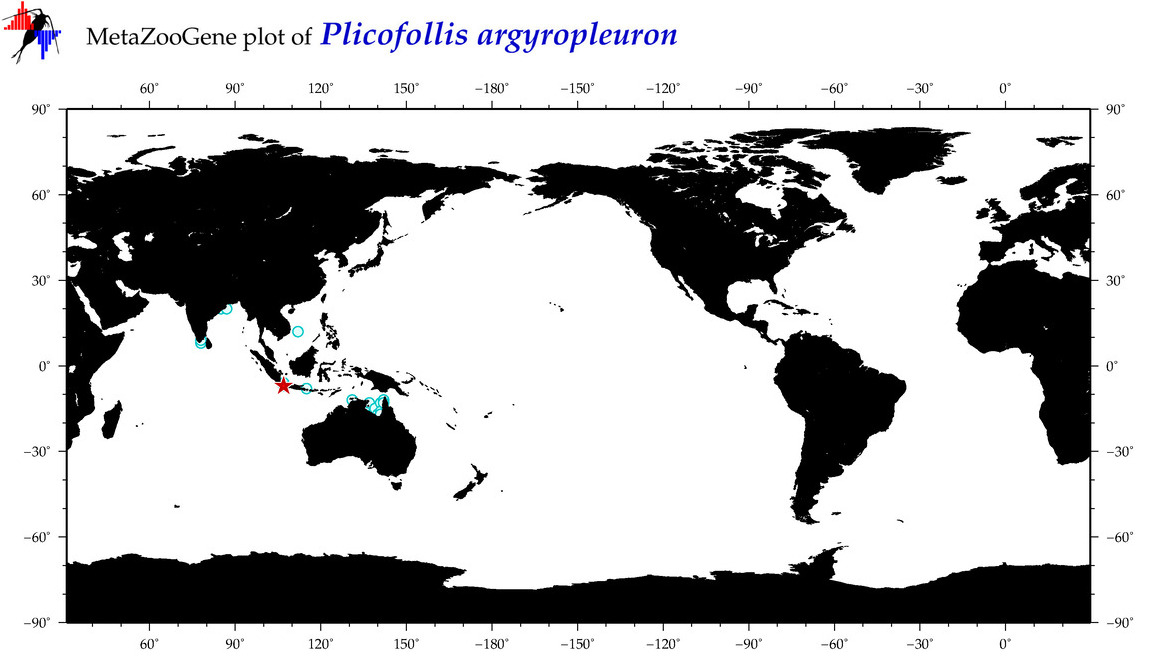 |
accepted
T5011468
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis dussumieri |
Species
(63) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
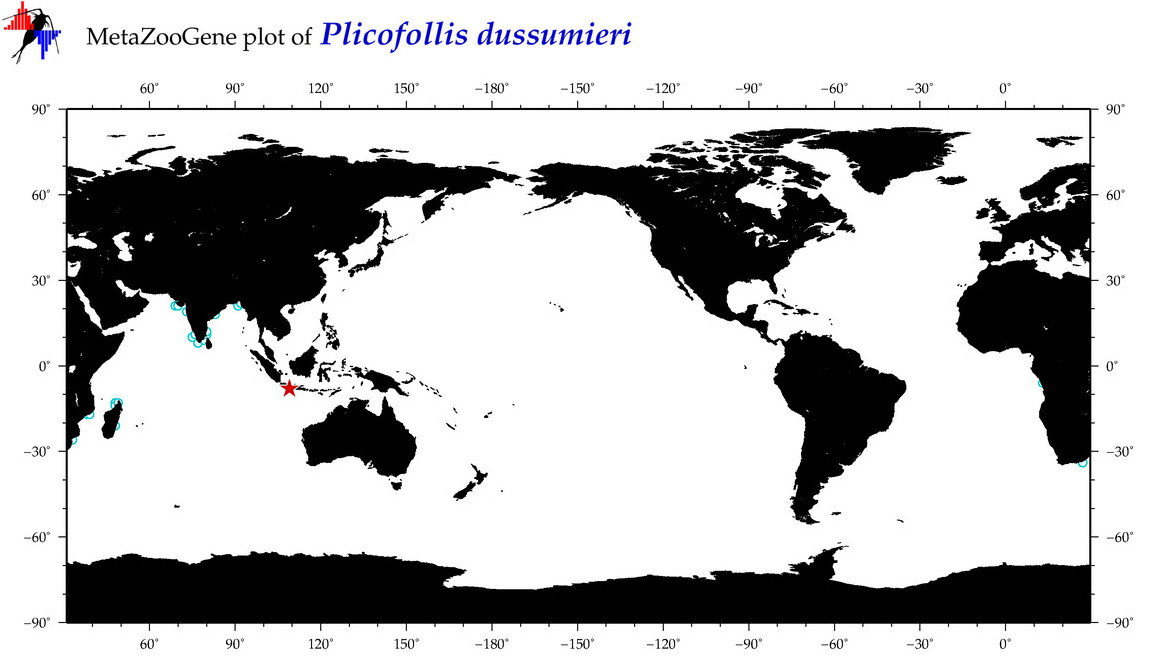 |
accepted
T5011469
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis layardi |
Species
(64) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 10
12S = 1
16S = 1
18S = 0
28S = 0
|
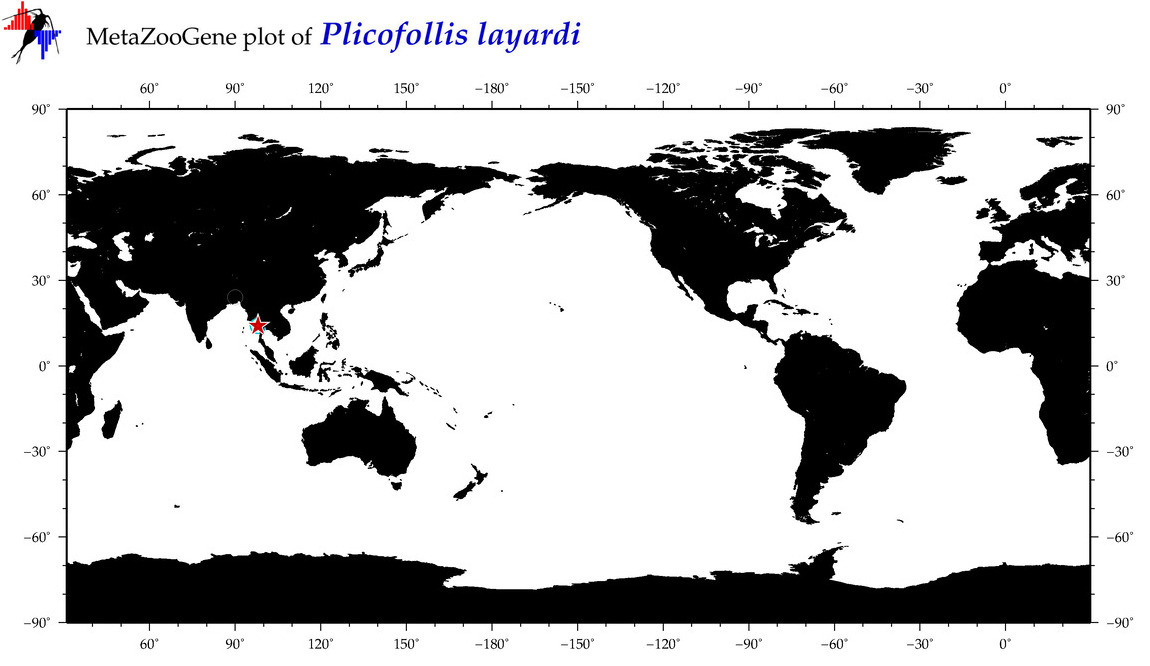 |
accepted
T5011470
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis nella |
Species
(65) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 5
12S = 2
16S = 1
18S = 0
28S = 0
|
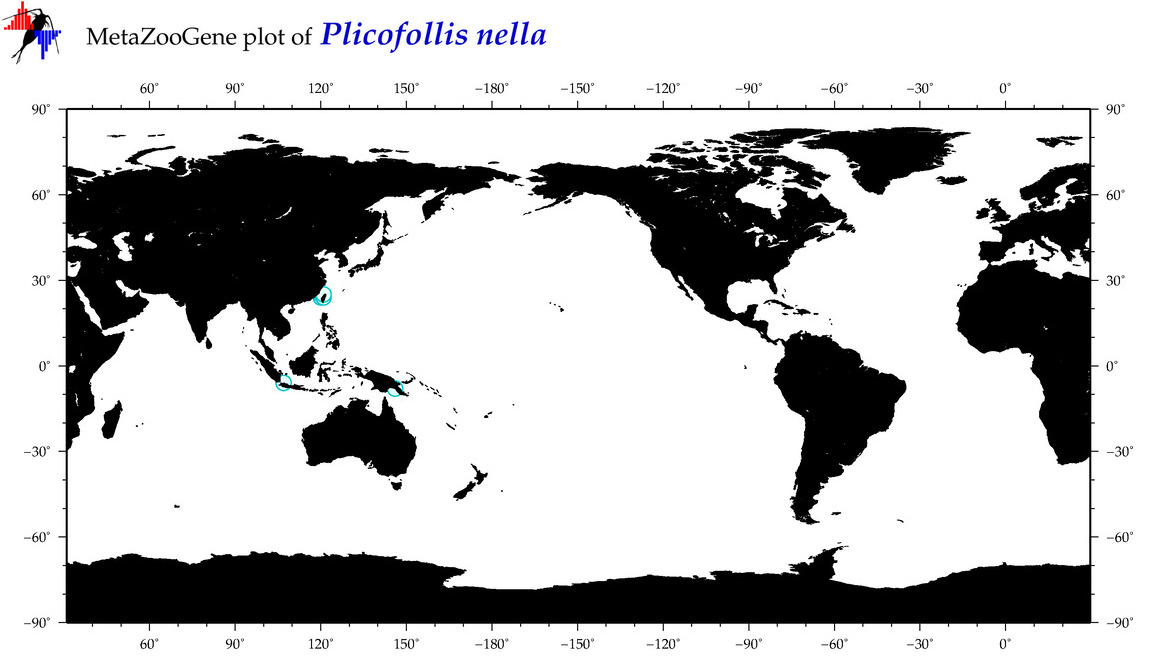 |
accepted
T5011472
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis platystomus |
Species
(66) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 1
16S = 0
18S = 0
28S = 0
|
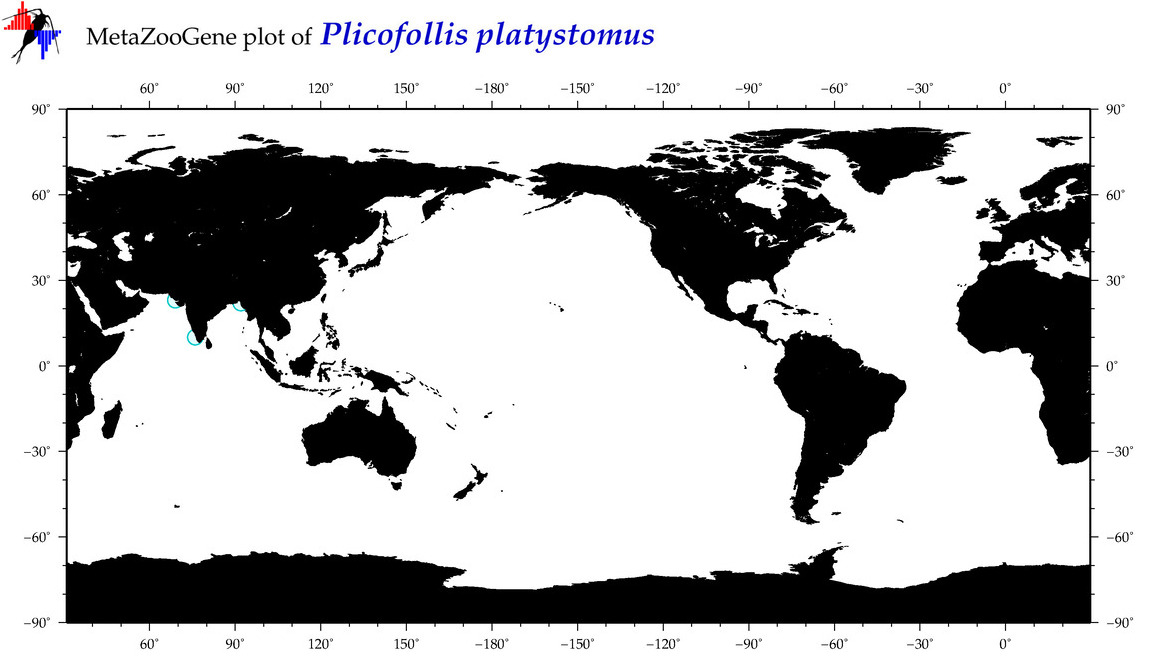 |
accepted
T5011473
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis polystaphylodon |
Species
(67) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 13
12S = 0
16S = 0
18S = 0
28S = 0
|
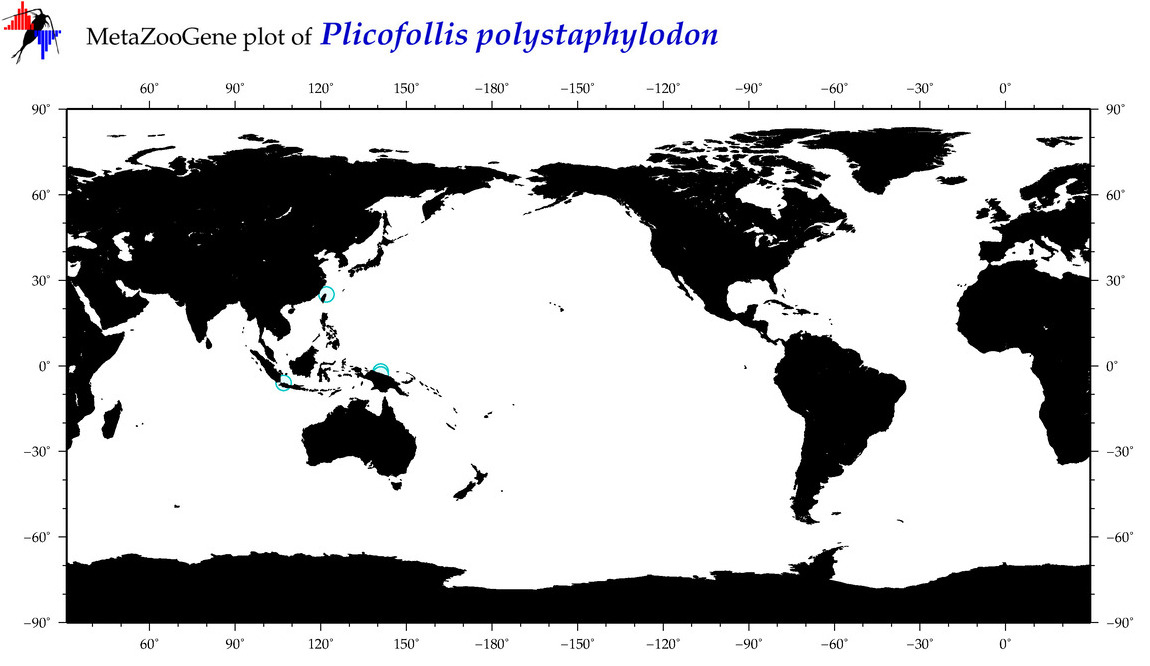 |
accepted
T5011474
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis tenuispinis |
Species
(68) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 16
12S = 0
16S = 0
18S = 0
28S = 0
|
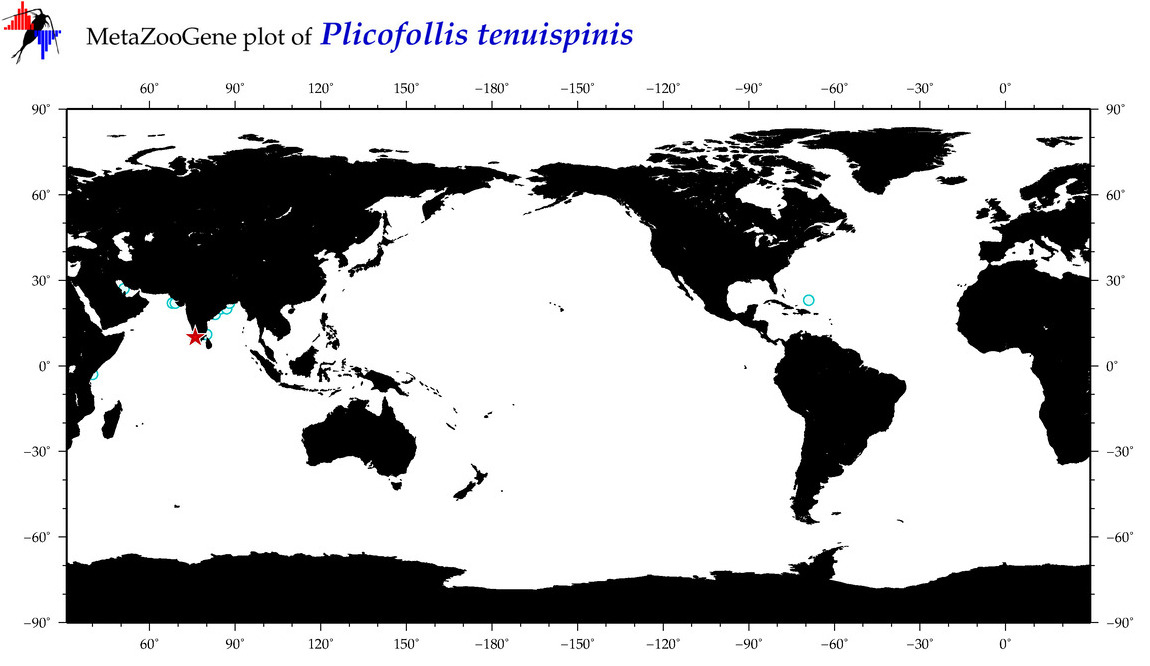 |
accepted
T5011475
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Plicofollis tonggol |
Species
(69) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5024137
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Potamarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5012865
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Sciades |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 7
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 8 |
Total Species: 7
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 8 |
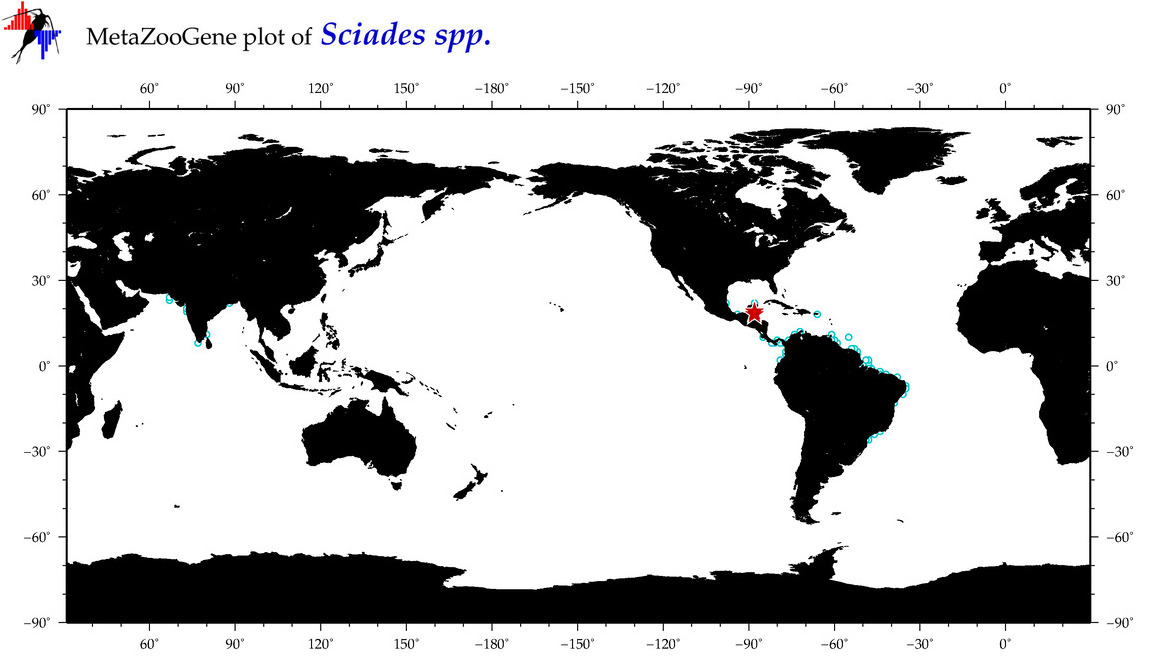 |
accepted
T5007303
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Sciades herzbergii |
Species
(70) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 1
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5025251
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Sciades parkeri |
Species
(71) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 2
18S = 0
28S = 0
|
no map |
accepted
T5025252
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Sciades passany |
Species
(72) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5025253
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Sciades proops |
Species
(73) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 2
18S = 0
28S = 0
|
no map |
accepted
T5025254
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Sciades sona |
Species
(74) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 7
12S = 1
16S = 1
18S = 0
28S = 0
|
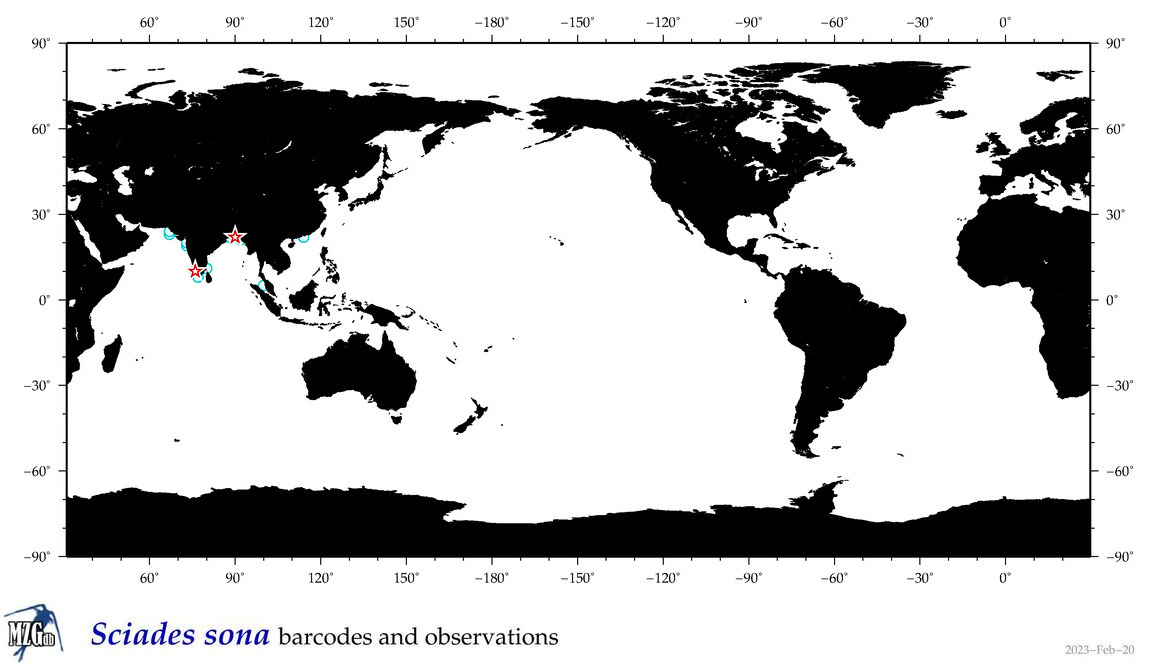 |
accepted
T5025255
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Ariopsis felis |
Species
(75) |
COI
12S
16S
18S
28S
|
COI = 29
12S = 1
16S = 1
18S = 0
28S = 1
|
COI = 29
12S = 1
16S = 1
18S = 0
28S = 1
|
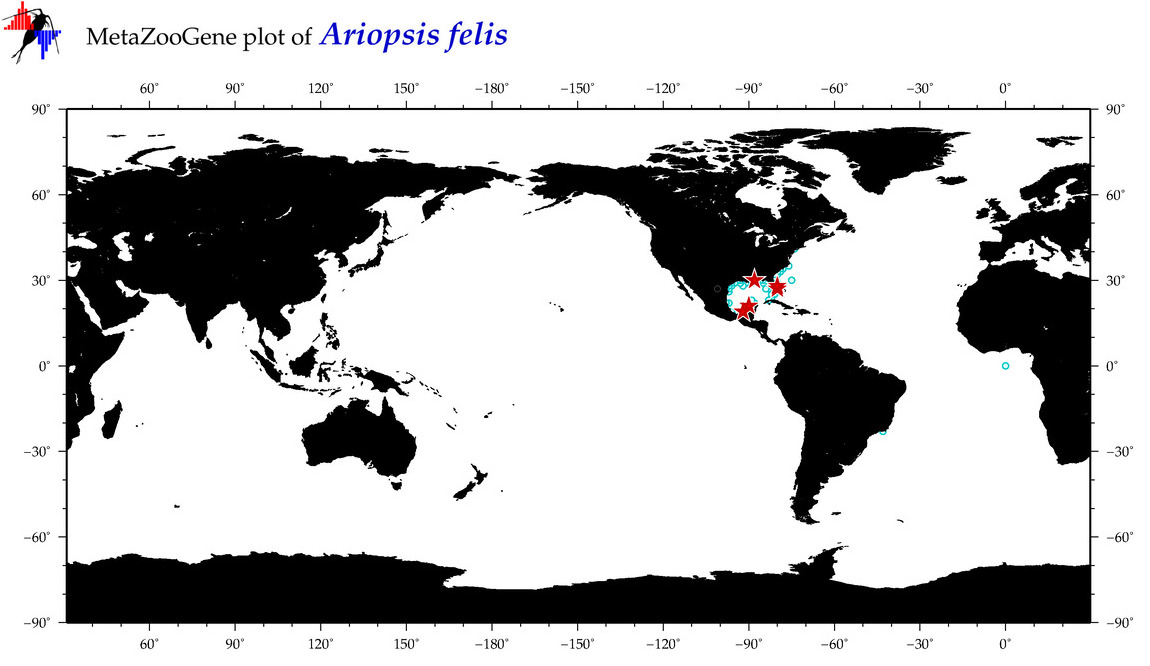 |
accepted
T5007889
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Ariopsis guatemalensis |
Species
(76) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5016614
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Ariopsis seemanni |
Species
(77) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 3
12S = 2
16S = 2
18S = 0
28S = 0
|
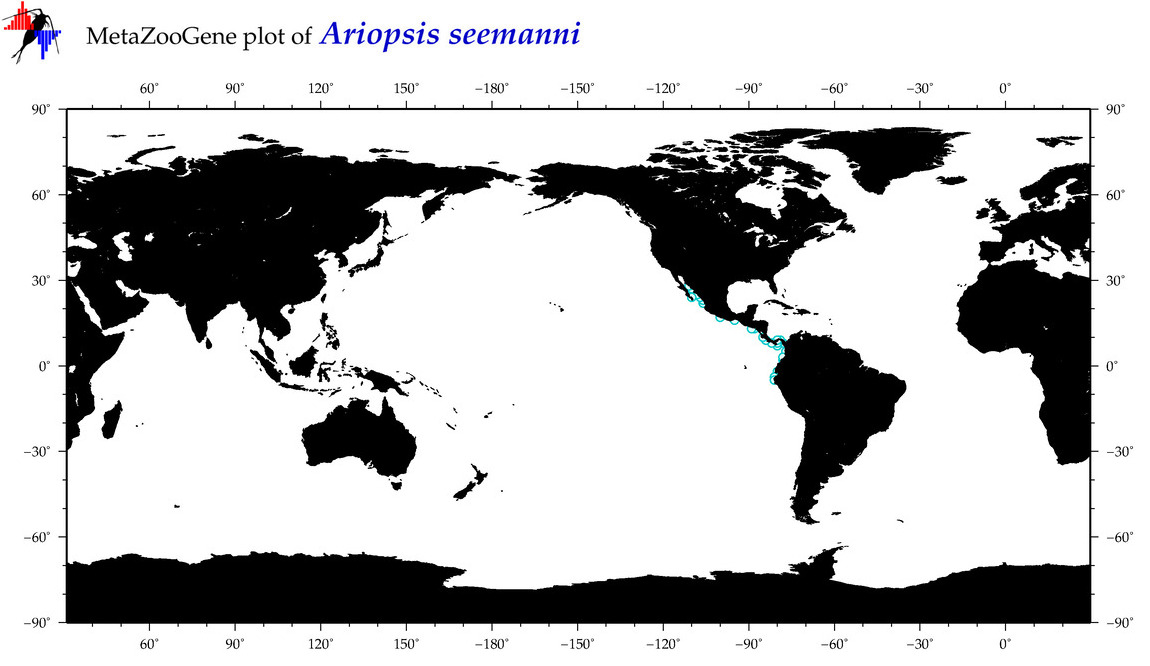 |
accepted
T5012972
R:1:1:1:0 |
| ⋄ ⋄ Bagreinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 134 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 134 |
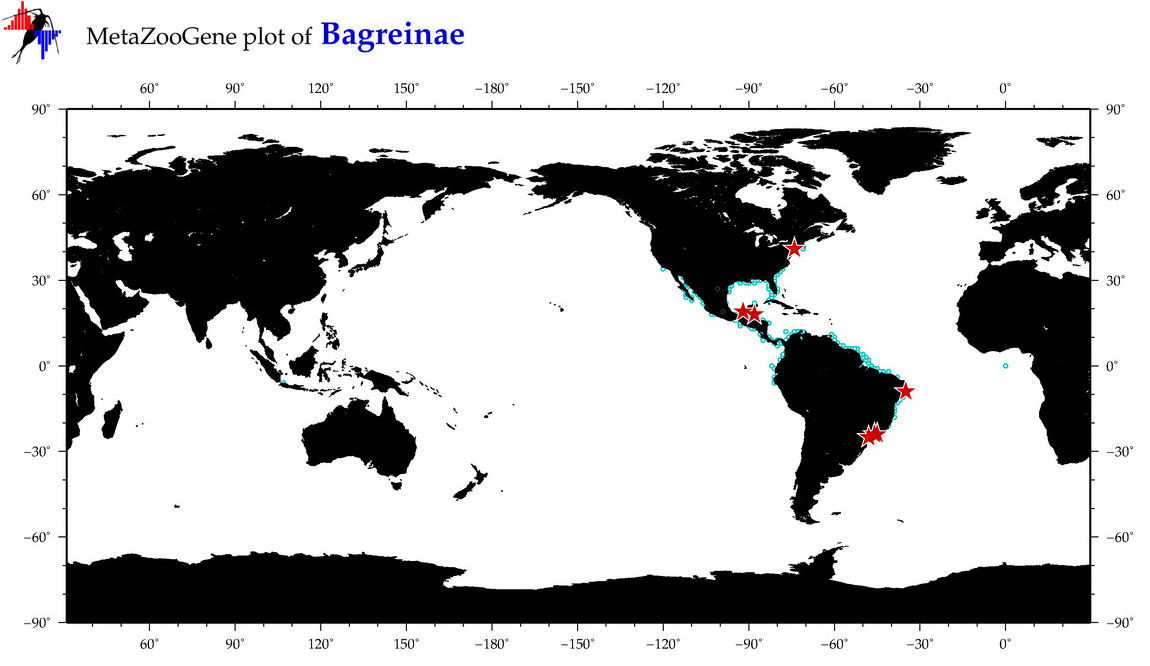 |
accepted
T5006197
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Bagre |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 134 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 134 |
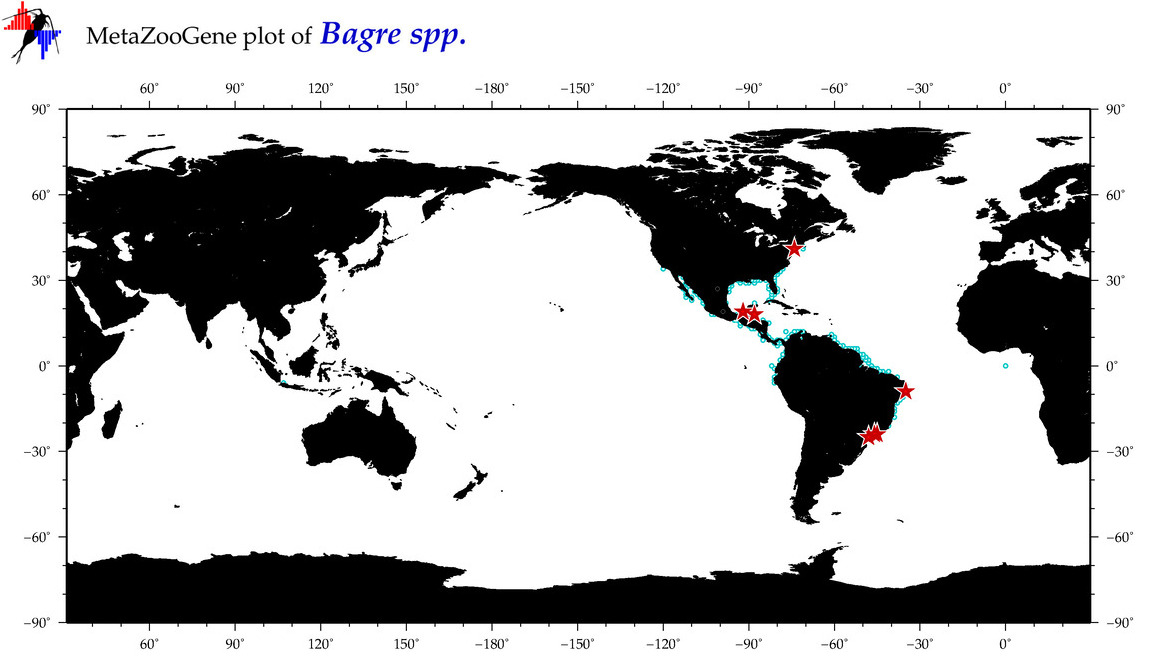 |
accepted
T5006292
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Bagre bagre |
Species
(78) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 14
12S = 1
16S = 1
18S = 0
28S = 0
|
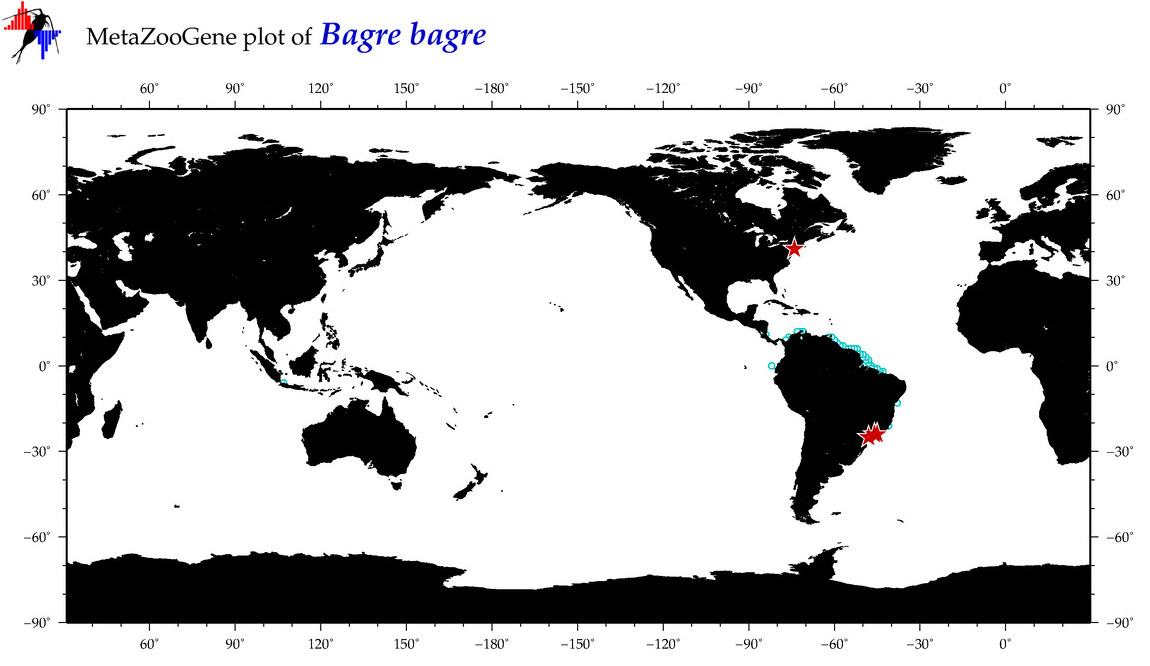 |
accepted
T5008036
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Bagre marinus |
Species
(79) |
COI
12S
16S
18S
28S
|
COI = 21
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 21
12S = 2
16S = 2
18S = 0
28S = 0
|
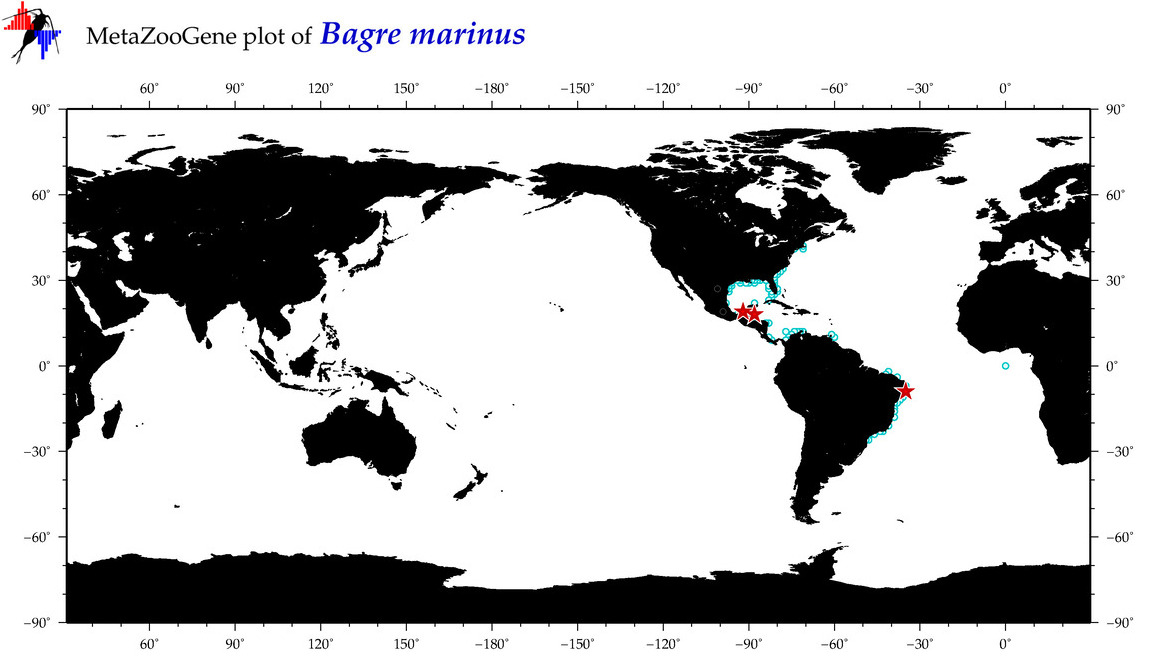 |
accepted
T5008037
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Bagre panamensis |
Species
(80) |
COI
12S
16S
18S
28S
|
COI = 99
12S = 3
16S = 3
18S = 0
28S = 0
|
COI = 99
12S = 3
16S = 3
18S = 0
28S = 0
|
no map |
accepted
T5016884
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Bagre pinnimaculatus |
Species
(81) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5016885
R:1:1:1:0 |
| ⋄ ⋄ Galeichthyinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
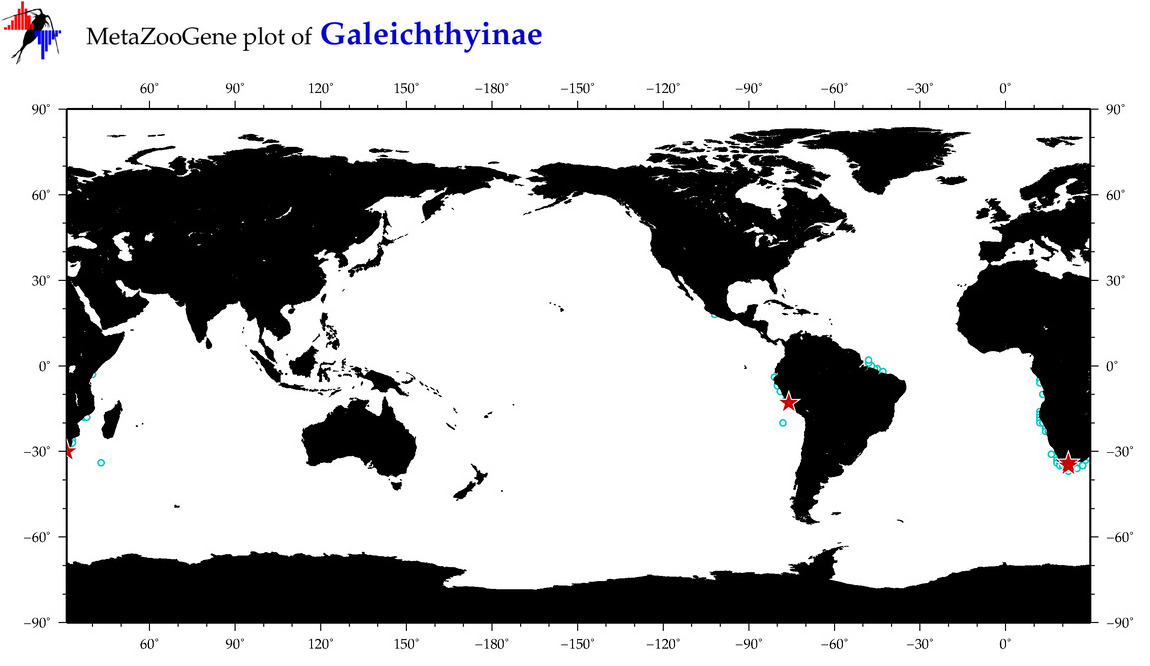 |
accepted
T5006198
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Galeichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
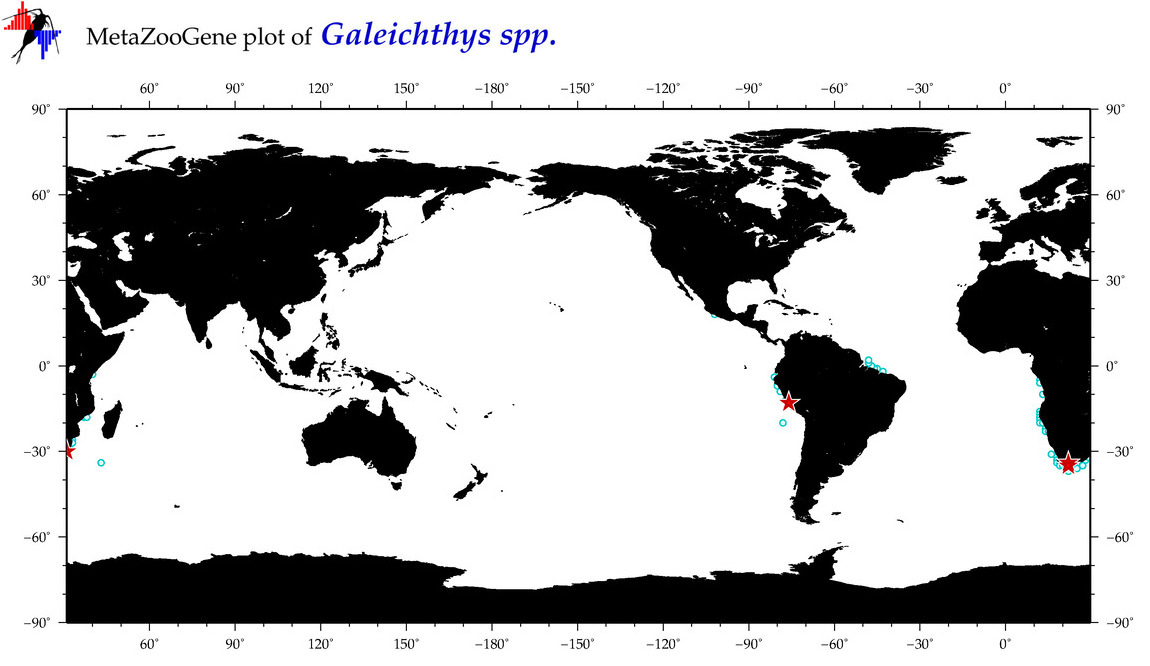 |
accepted
T5006318
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Galeichthys ater |
Species
(82) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 4
12S = 1
16S = 1
18S = 0
28S = 0
|
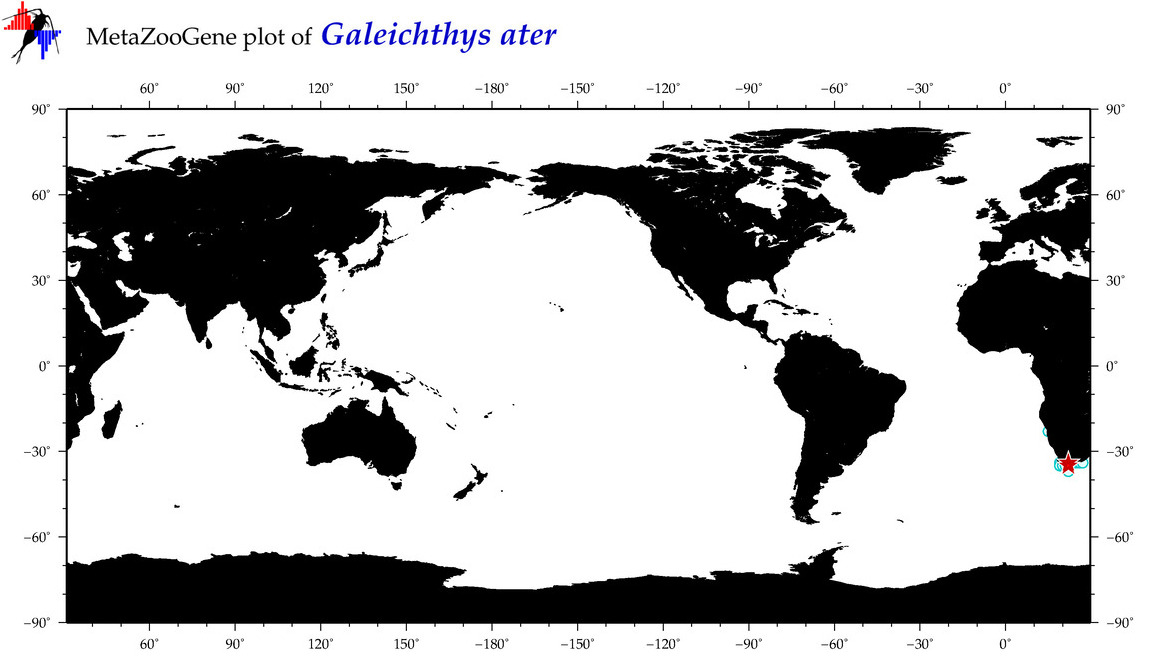 |
accepted
T5009252
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Galeichthys feliceps |
Species
(83) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
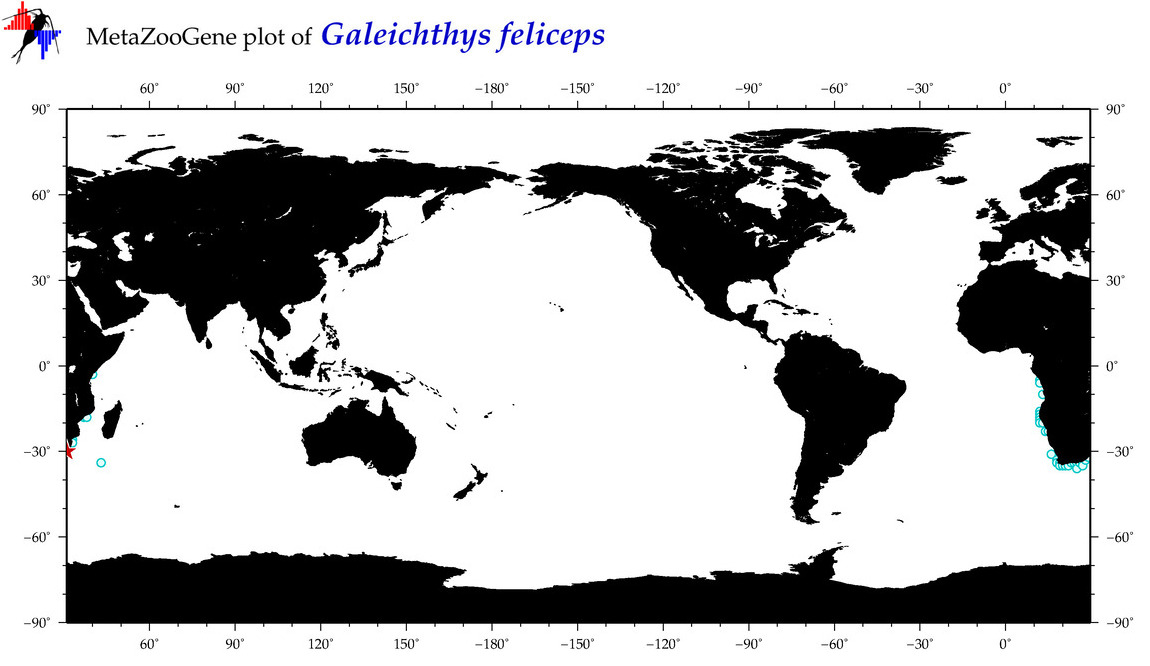 |
accepted
T5009253
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Galeichthys peruvianus |
Species
(84) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
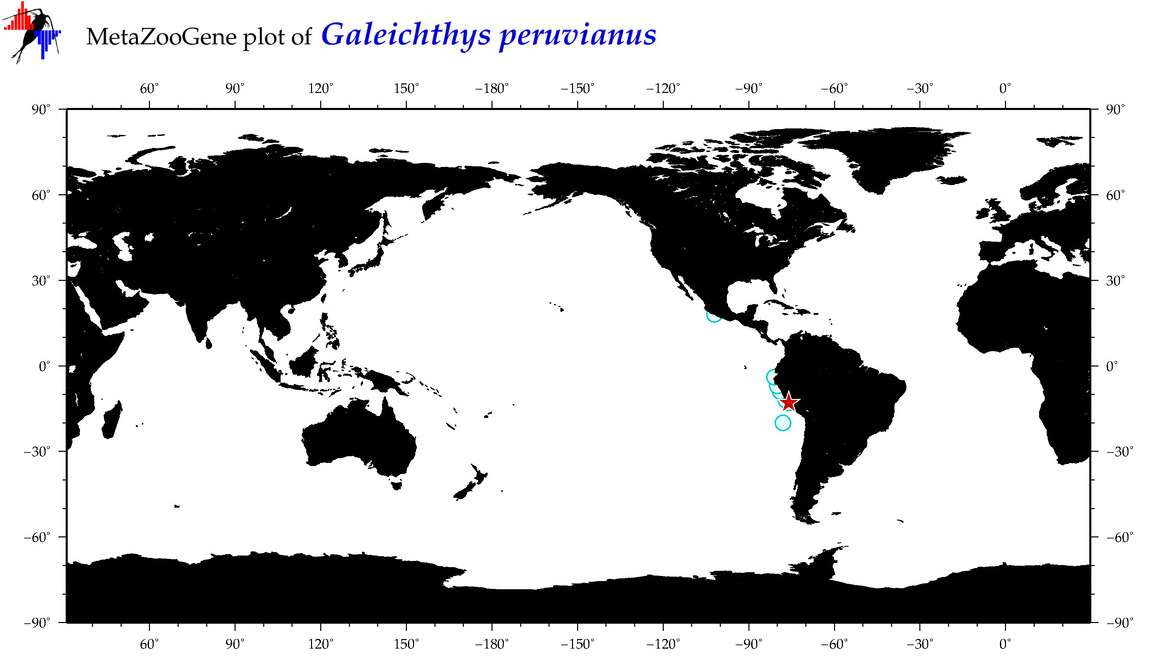 |
accepted
T5009254
R:1:0:0:0 |
| ⋄ Aspredinidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 9 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 9 |
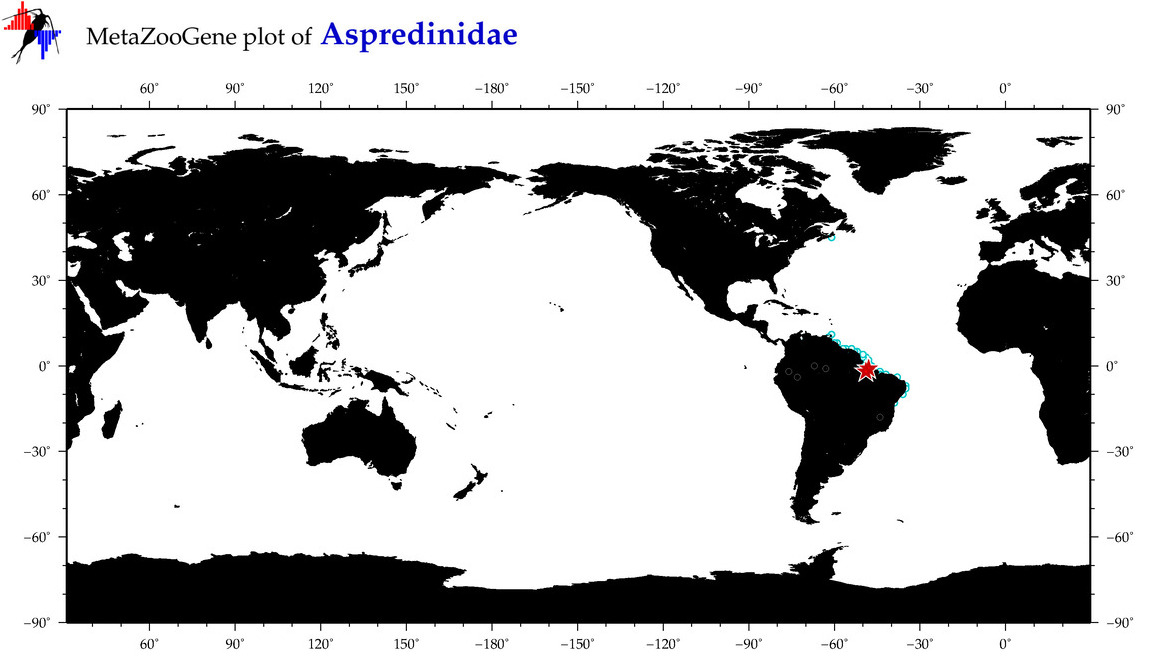 |
accepted
T5005961
R:0:1:1:0 |
| ⋄ ⋄ Aspredinichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
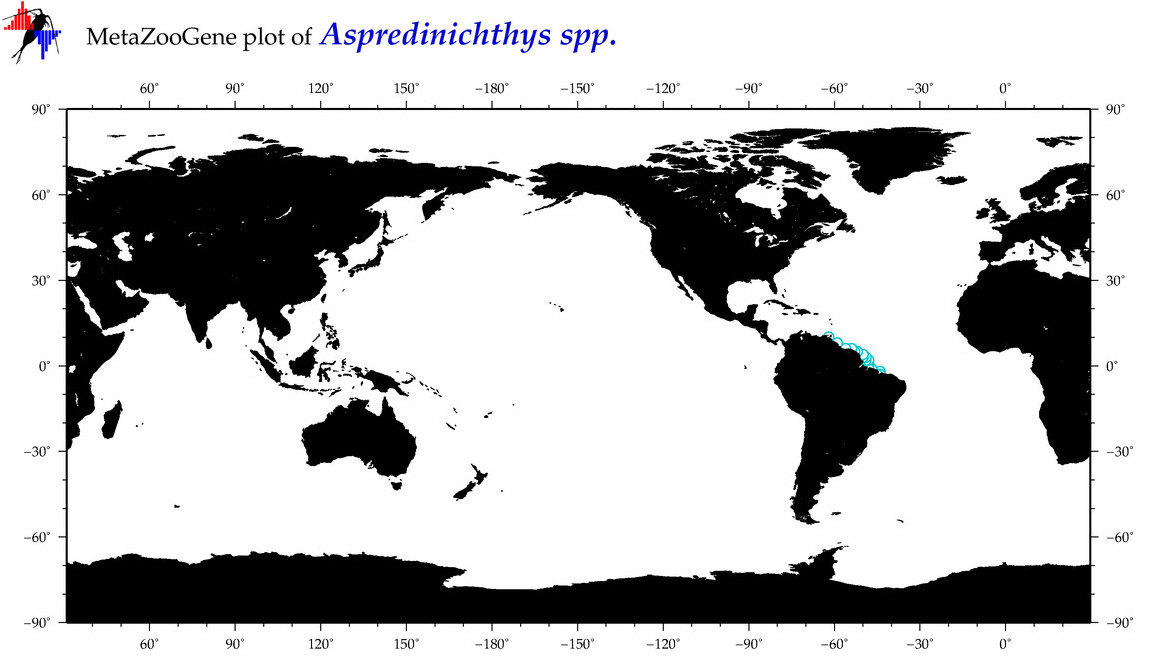 |
accepted
T5006489
R:0:1:1:0 |
| ⋄ ⋄ Aspredo |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
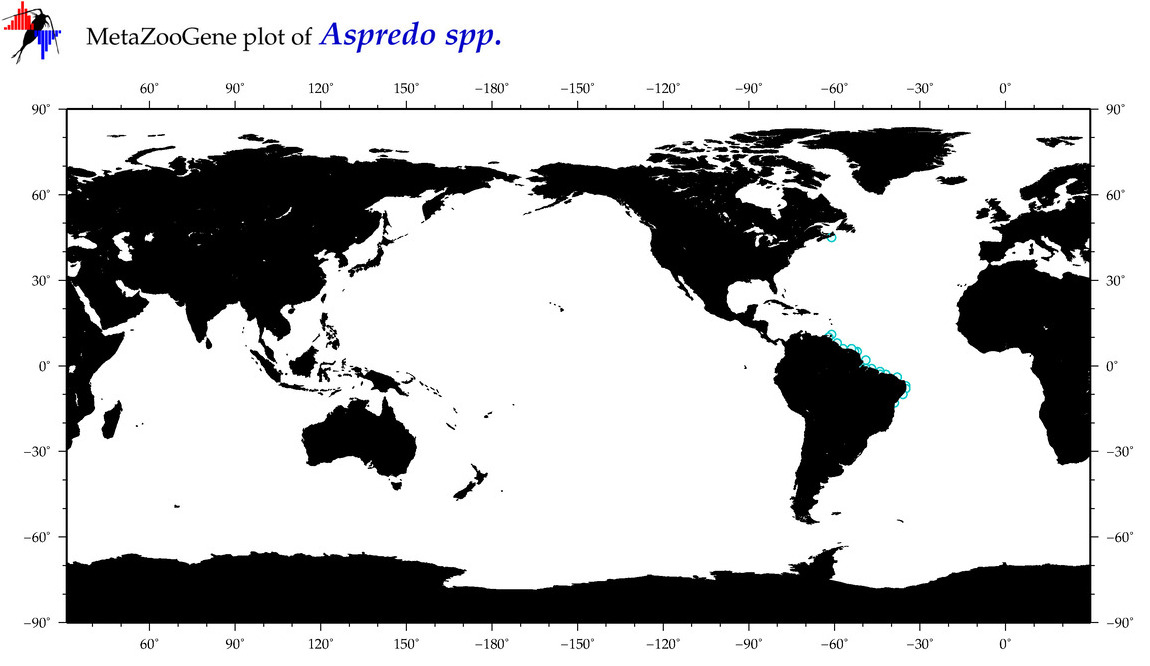 |
accepted
T5006490
R:0:1:1:0 |
| ⋄ ⋄ Platystacus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
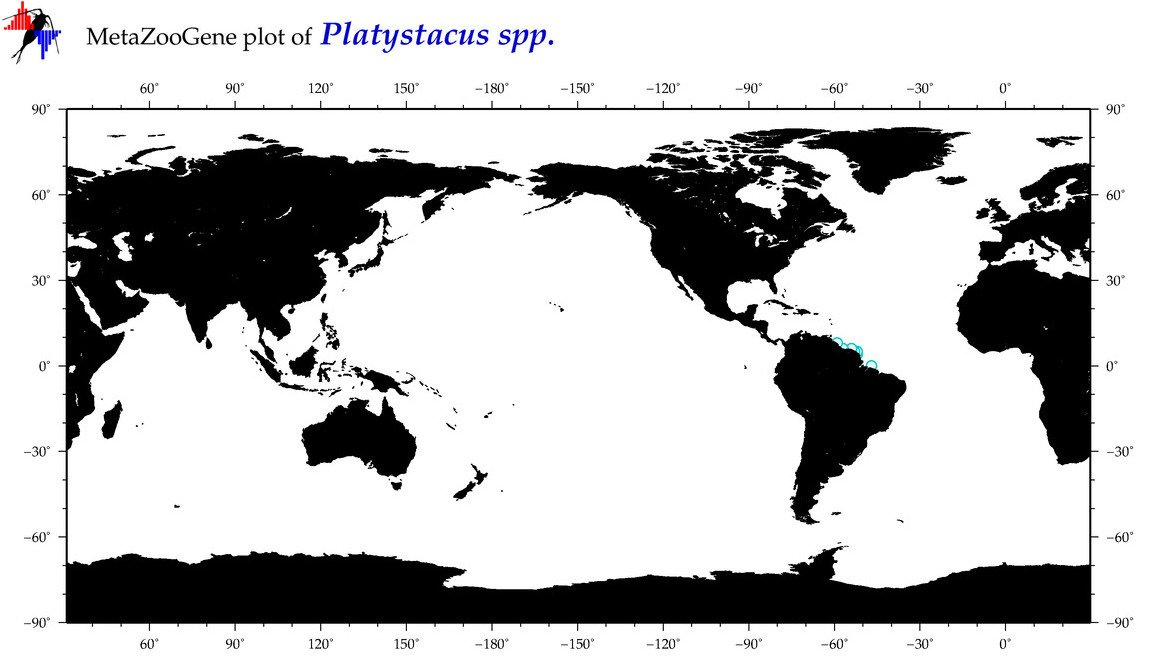 |
accepted
T5007156
R:0:1:1:0 |
| ⋄ Auchenipteridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
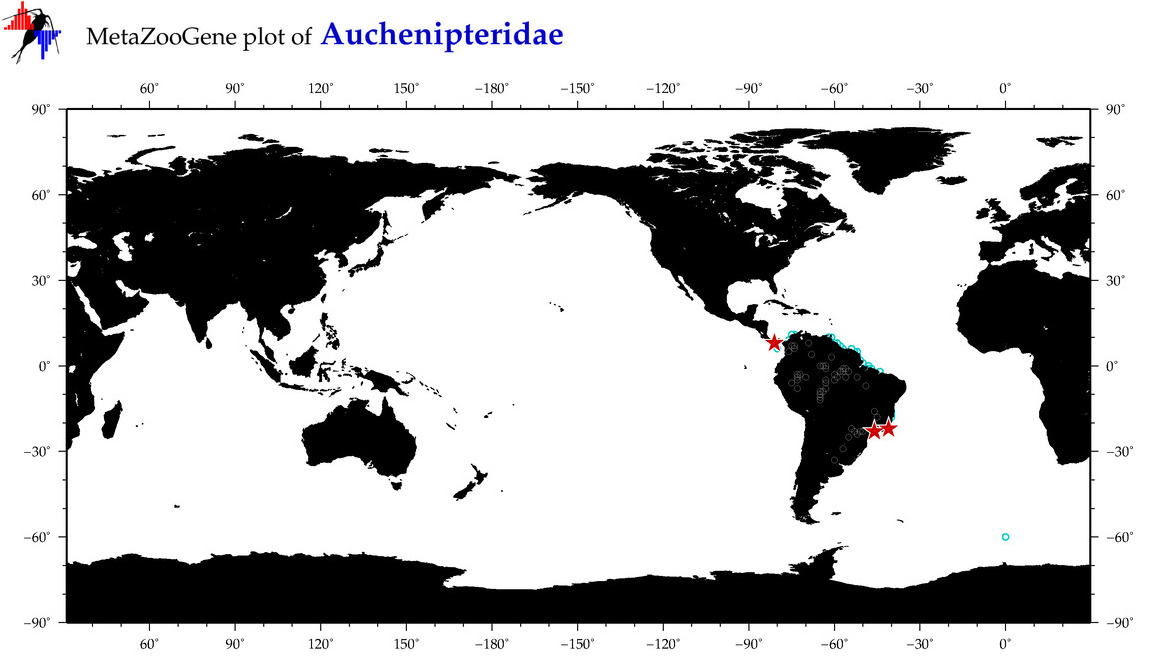 |
accepted
T5005995
R:0:1:1:0 |
| ⋄ ⋄ Auchenipterinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
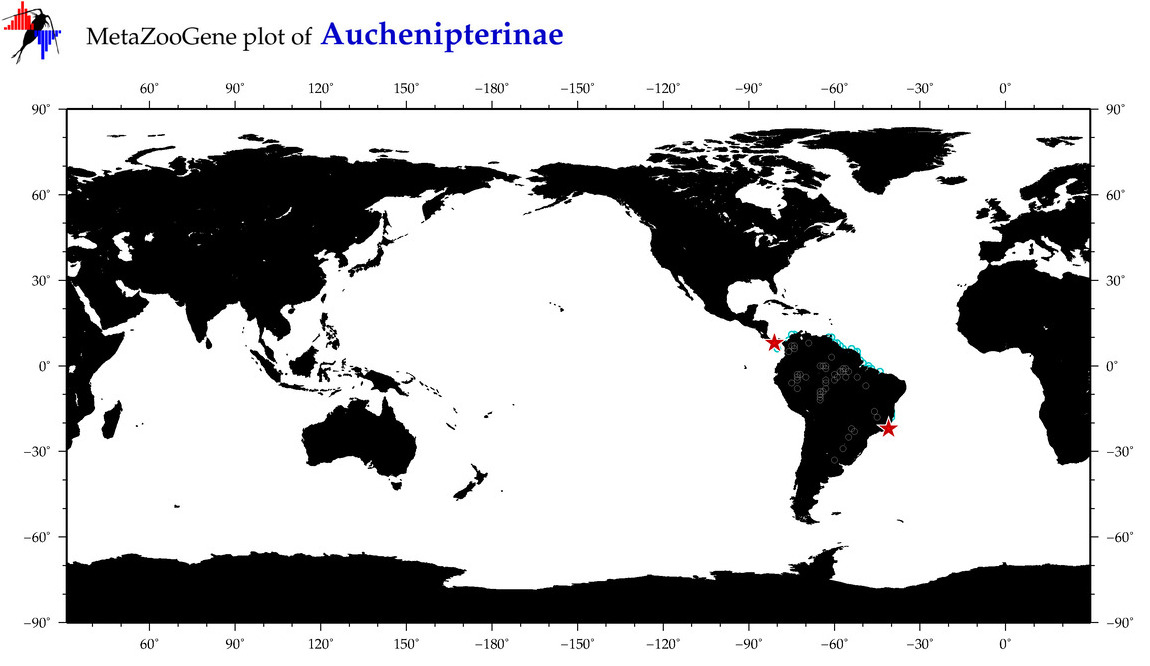 |
accepted
T5006133
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Pseudauchenipterus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
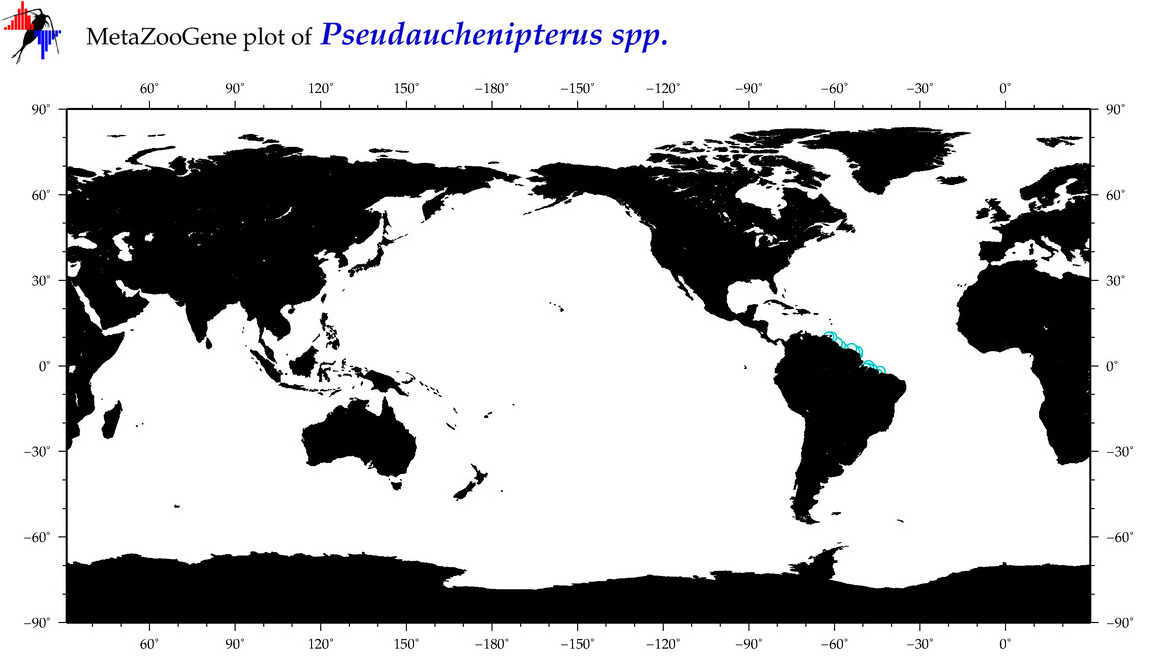 |
accepted
T5007189
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Tetranematichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5007369
R:0:1:1:0 |
| ⋄ Bagridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 19
Sp. w/COI-Barcodes: 17
Total COI-Barcodes: 703 |
Total Species: 19
Sp. w/COI-Barcodes: 17
Total COI-Barcodes: 703 |
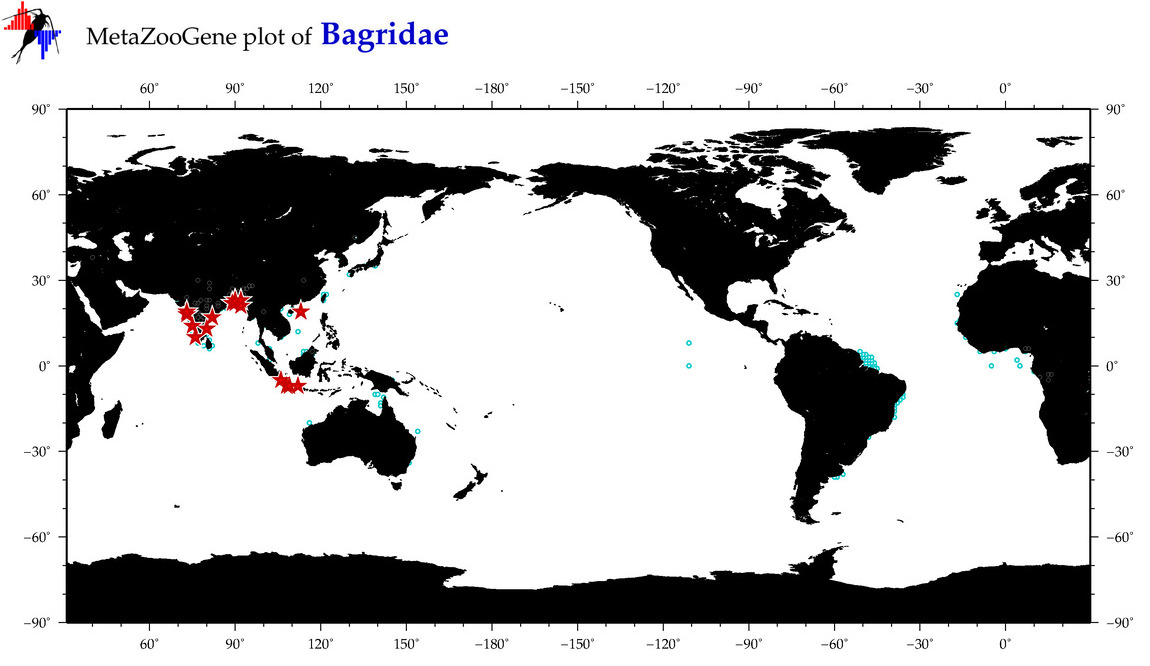 |
accepted
T5006016
R:0:1:1:0 |
| ⋄ ⋄ Claroteinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 73 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 73 |
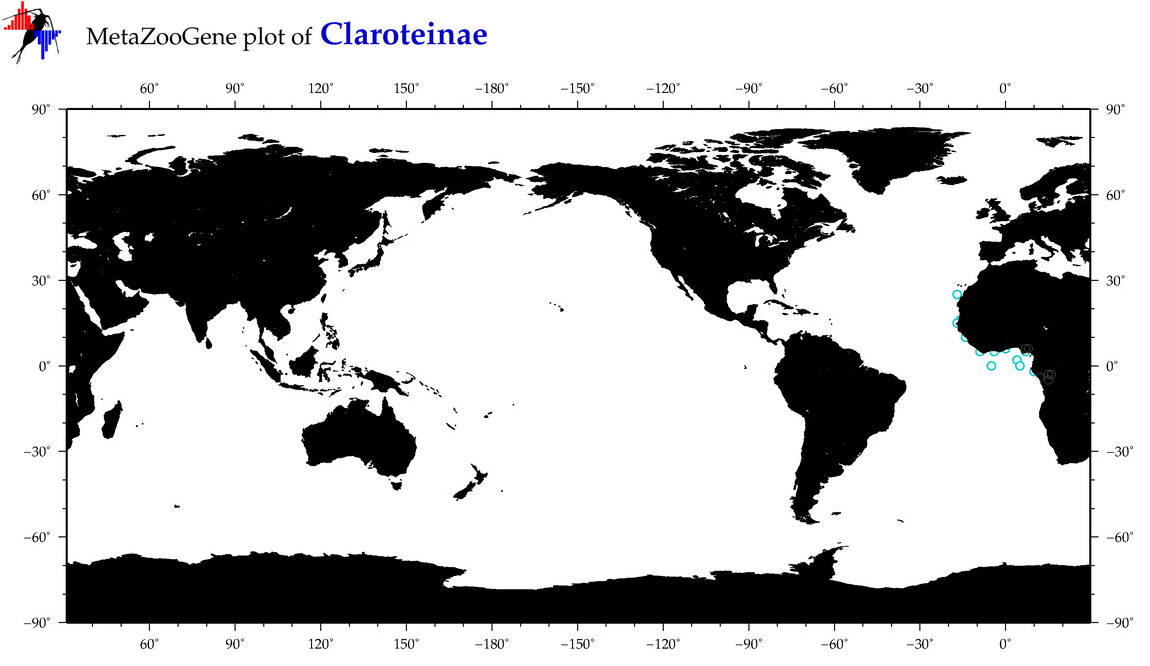 |
accepted
T5006134
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Chrysichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 73 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 73 |
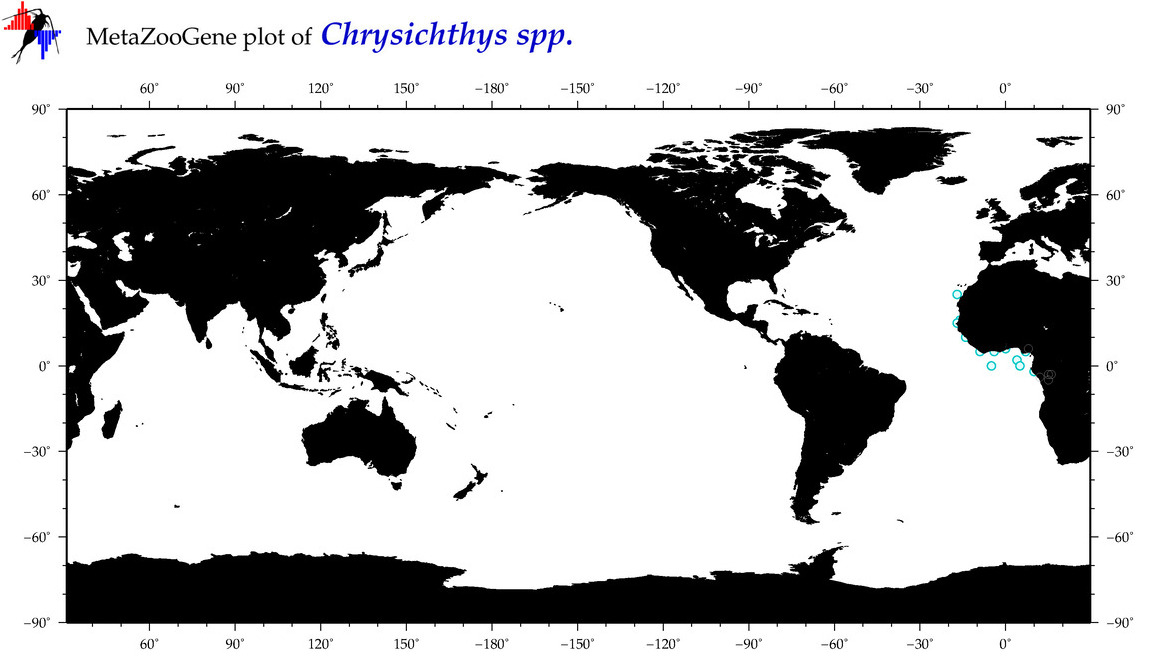 |
accepted
T5006613
R:0:1:1:0 |
| ⋄ ⋄ Hemibagrus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 117 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 117 |
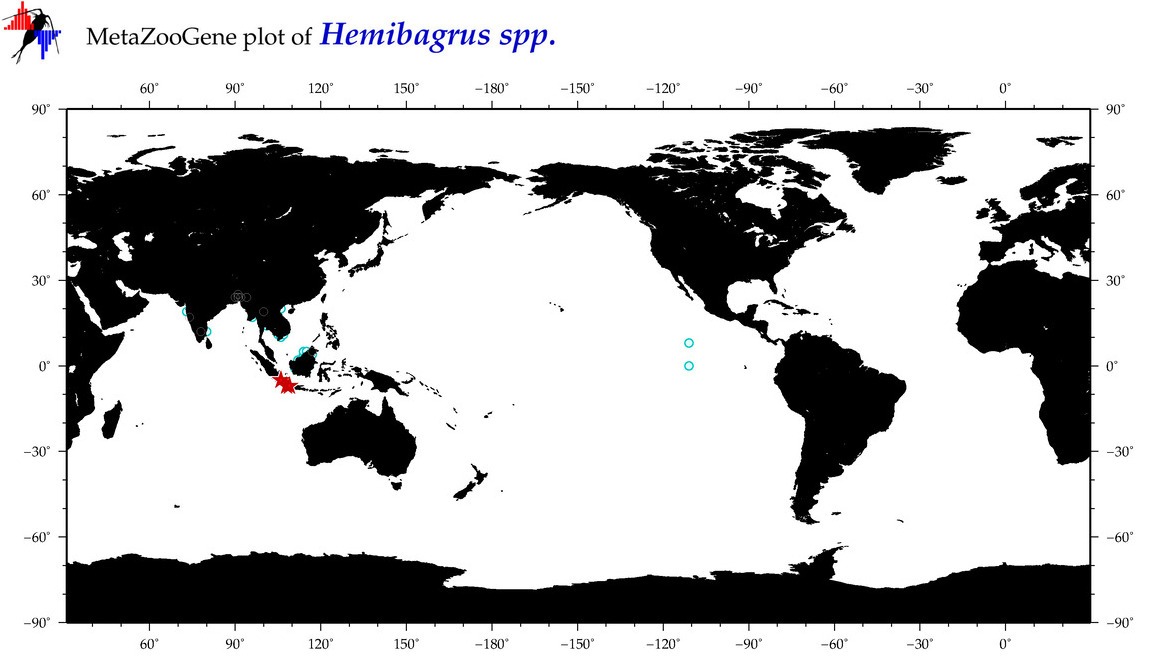 |
accepted
T5006778
R:0:1:1:0 |
| ⋄ ⋄ Mystus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 10
Sp. w/COI-Barcodes: 9
Total COI-Barcodes: 291 |
Total Species: 10
Sp. w/COI-Barcodes: 9
Total COI-Barcodes: 291 |
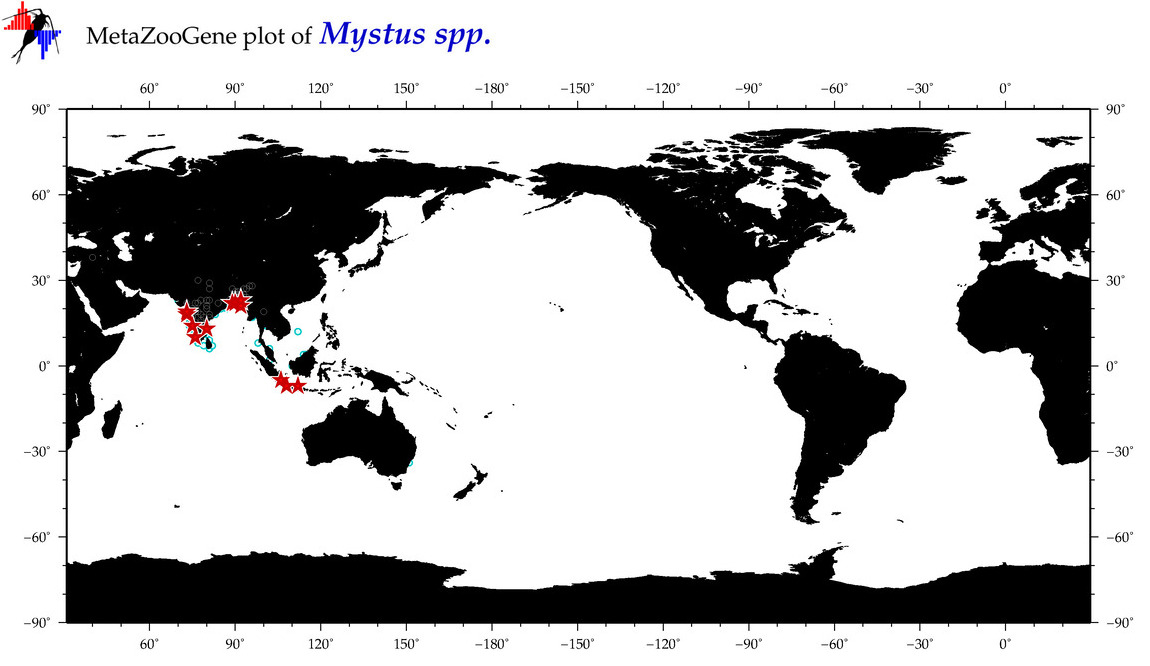 |
accepted
T5006981
R:0:1:1:1 |
| ⋄ ⋄ Pelteobagrus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
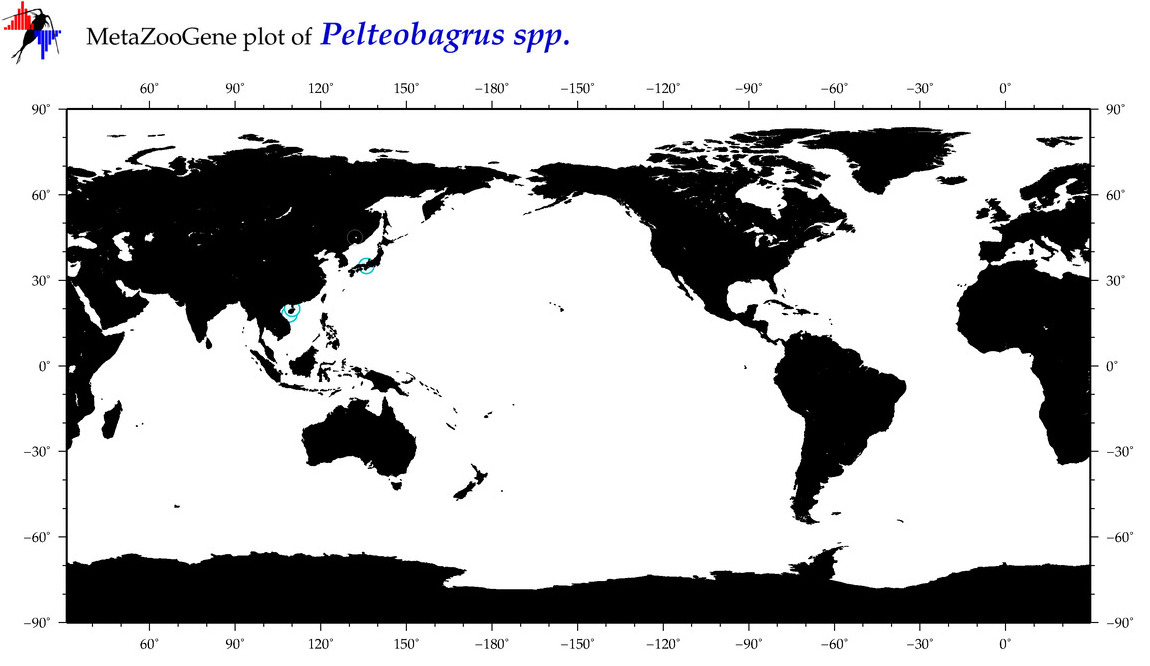 |
accepted
T5011261
R:0:1:1:1 |
| ⋄ ⋄ Rita |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 50 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 50 |
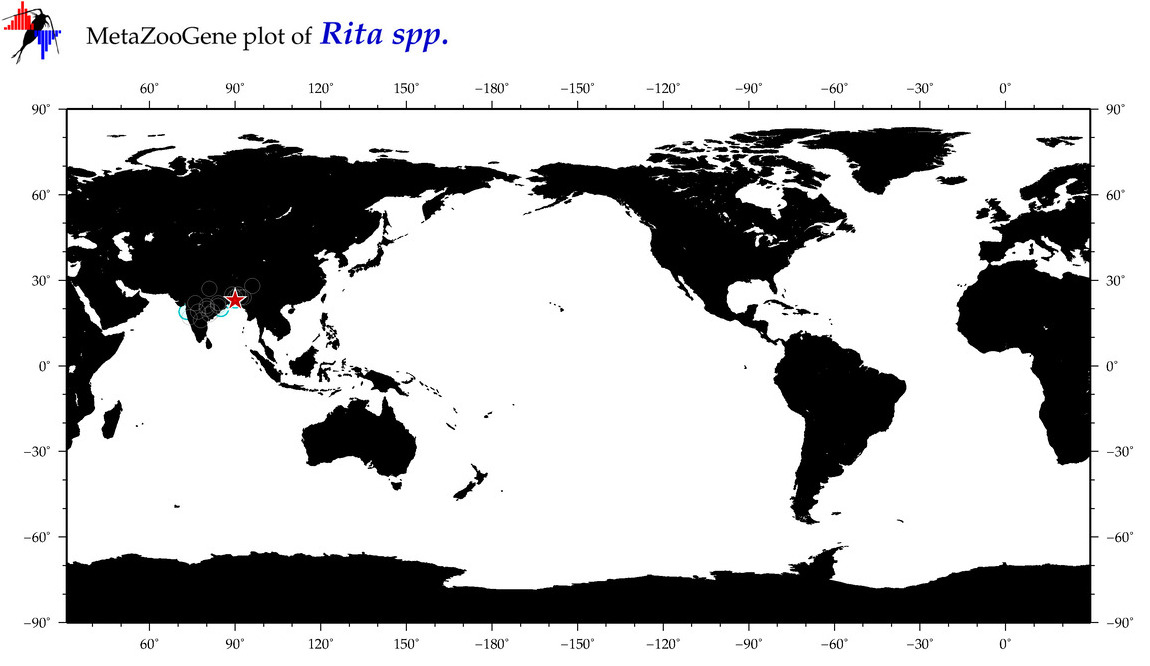 |
accepted
T5007273
R:0:1:1:0 |
| ⋄ ⋄ Sperata |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 102 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 102 |
 |
accepted
T5007334
R:0:1:1:0 |
| ⋄ ⋄ Tachysurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 70 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 70 |
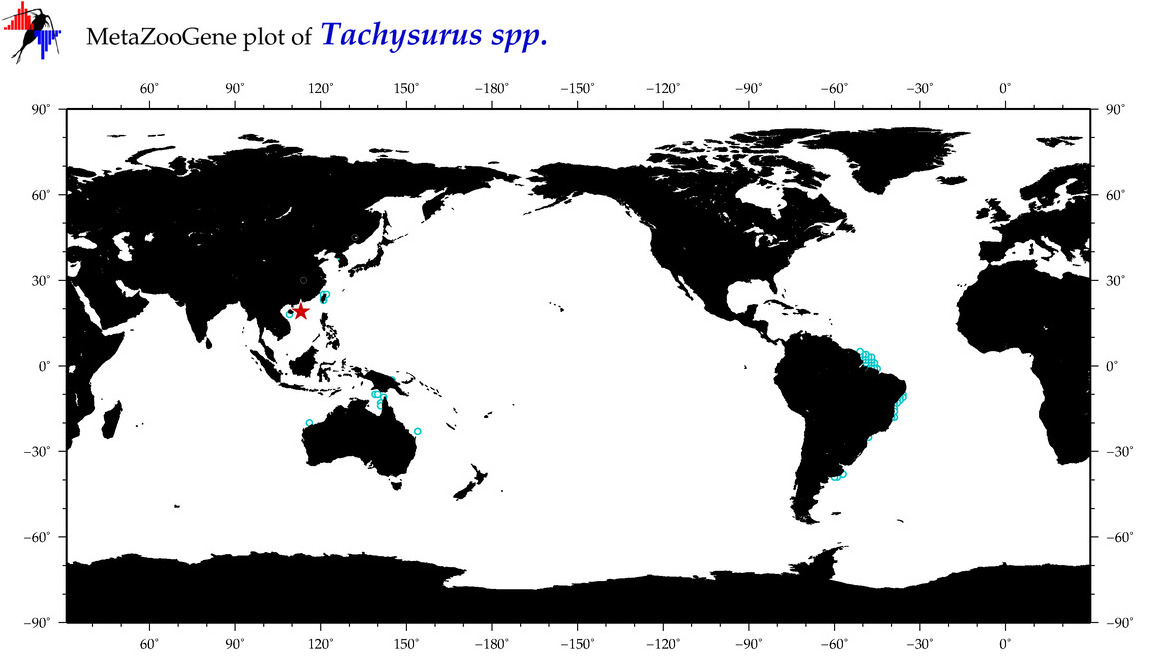 |
accepted
T5006328
R:0:1:1:0 |
| ⋄ Clariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 316 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 316 |
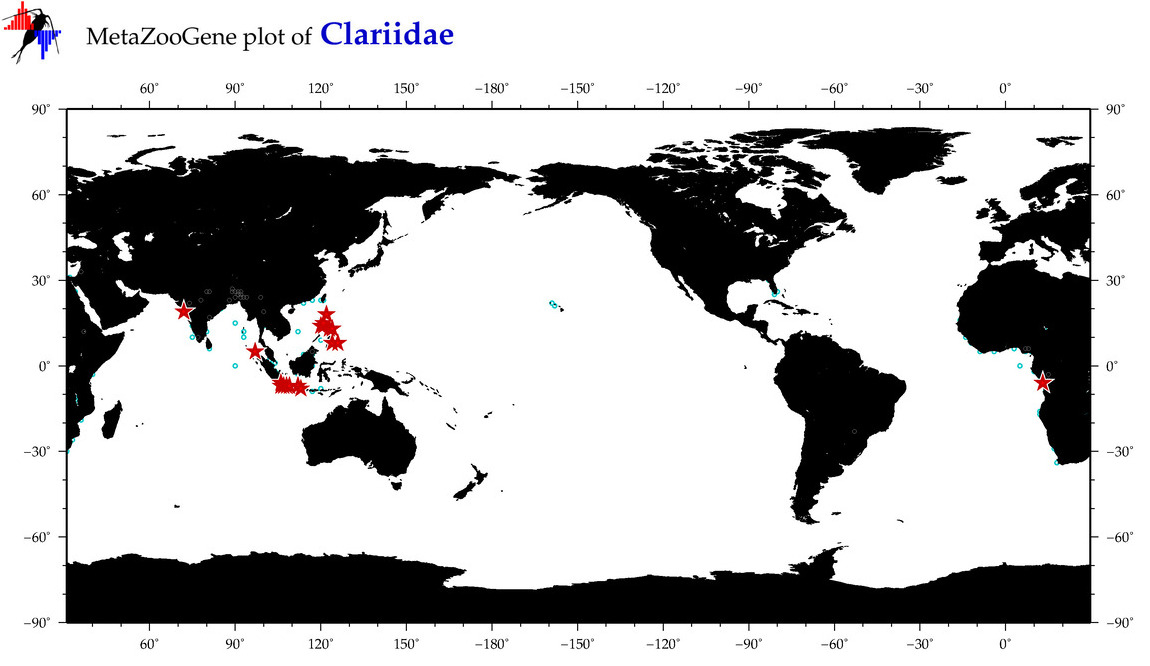 |
accepted
T5006025
R:0:1:1:0 |
| ⋄ ⋄ Clarias |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 316 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 316 |
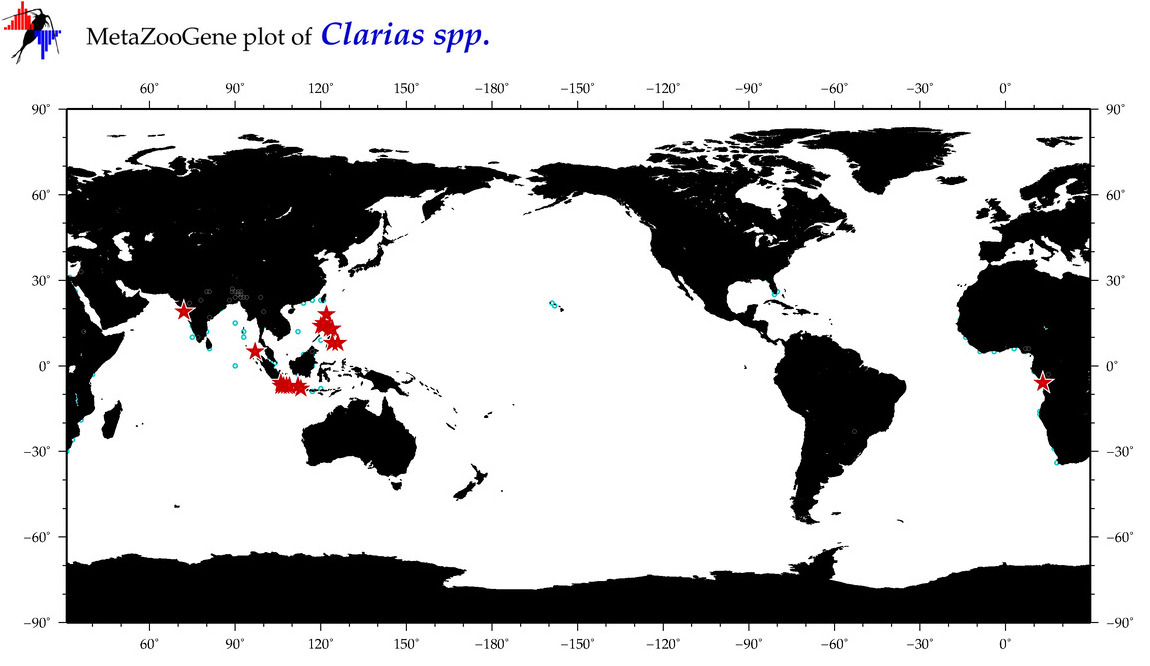 |
accepted
T5006619
R:0:1:1:0 |
| ⋄ Claroteidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006055
R:1:1:1:0 |
| ⋄ ⋄ Auchenoglanidinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006199
R:1:0:1:0 |
| ⋄ Doradidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5005979
R:0:1:1:0 |
| ⋄ ⋄ Doras |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006686
R:0:1:1:0 |
| ⋄ Heptapteridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 25 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 25 |
 |
accepted
T5006049
R:1:1:1:0 |
| ⋄ ⋄ Phreatobius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5029961
R:0:1:1:0 |
| ⋄ ⋄ Pimelodella |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5007138
R:0:1:1:0 |
| ⋄ ⋄ Rhamdia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 25 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 25 |
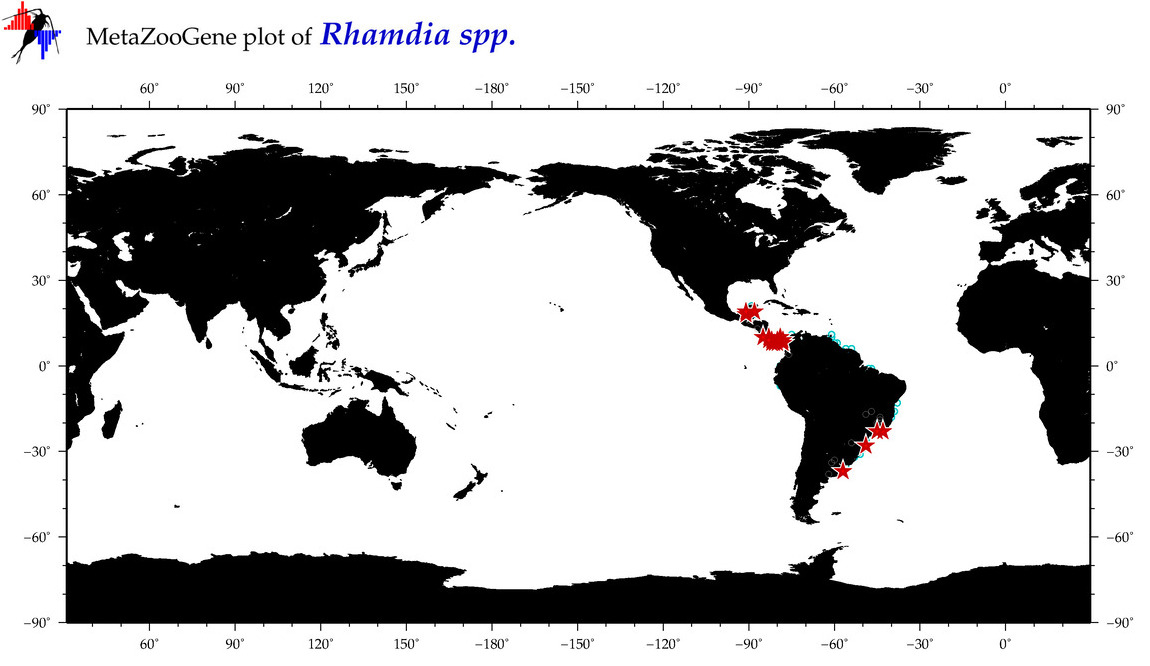 |
accepted
T5007257
R:1:0:1:0 |
| ⋄ ⋄ ⋄ Rhamdia eurycephala |
Species
(85) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5029733
R:1:0:1:0 |
| ⋄ ⋄ ⋄ Rhamdia gabrielae |
Species
(86) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 20
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5029734
R:1:0:1:0 |
| ⋄ Heteropneustidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 109 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 109 |
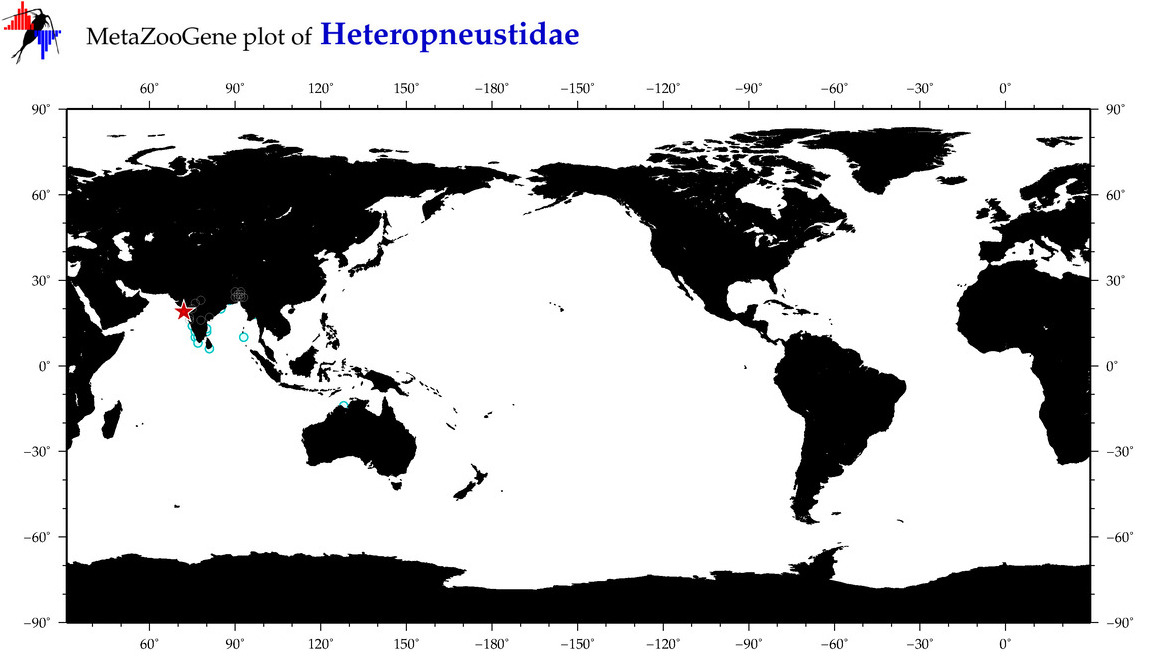 |
accepted
T5006026
R:0:1:1:0 |
| ⋄ ⋄ Heteropneustes |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 109 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 109 |
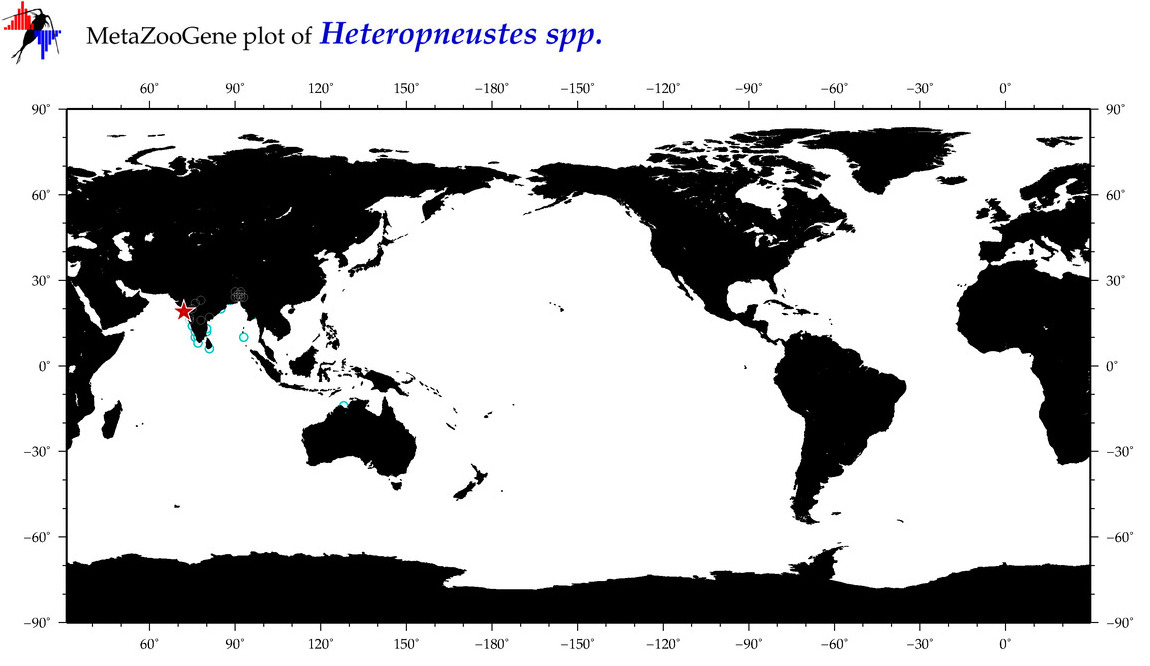 |
accepted
T5006803
R:0:1:1:0 |
| ⋄ Horabagridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 23 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 23 |
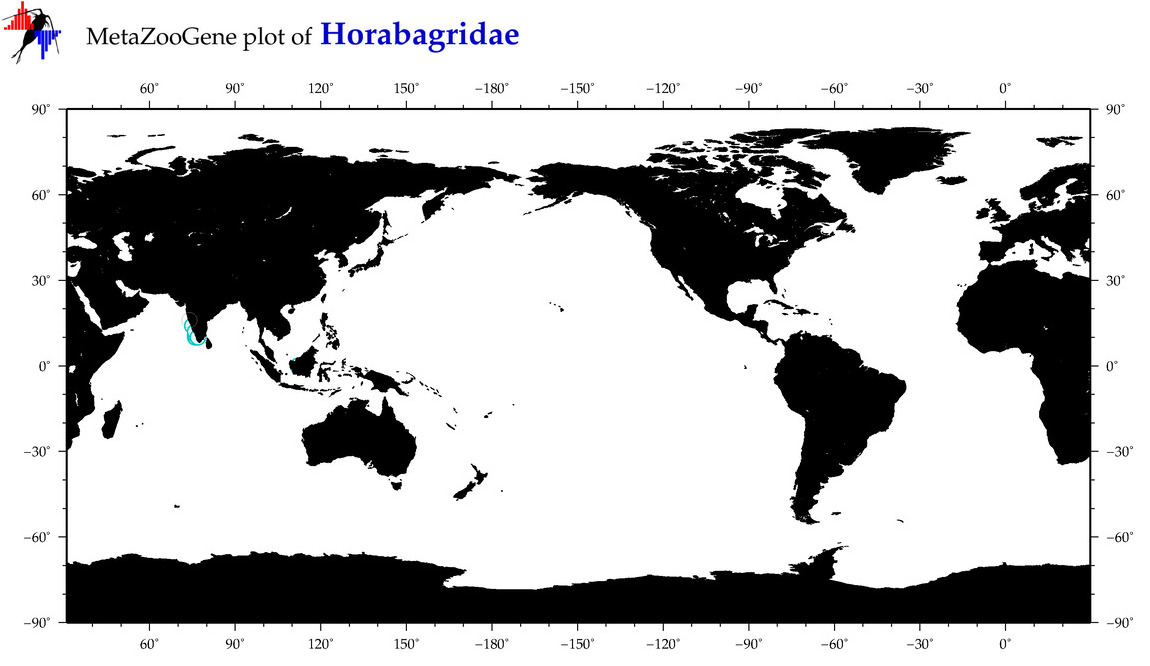 |
accepted
T5006062
R:0:1:1:0 |
| ⋄ ⋄ Horabagrus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 23 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 23 |
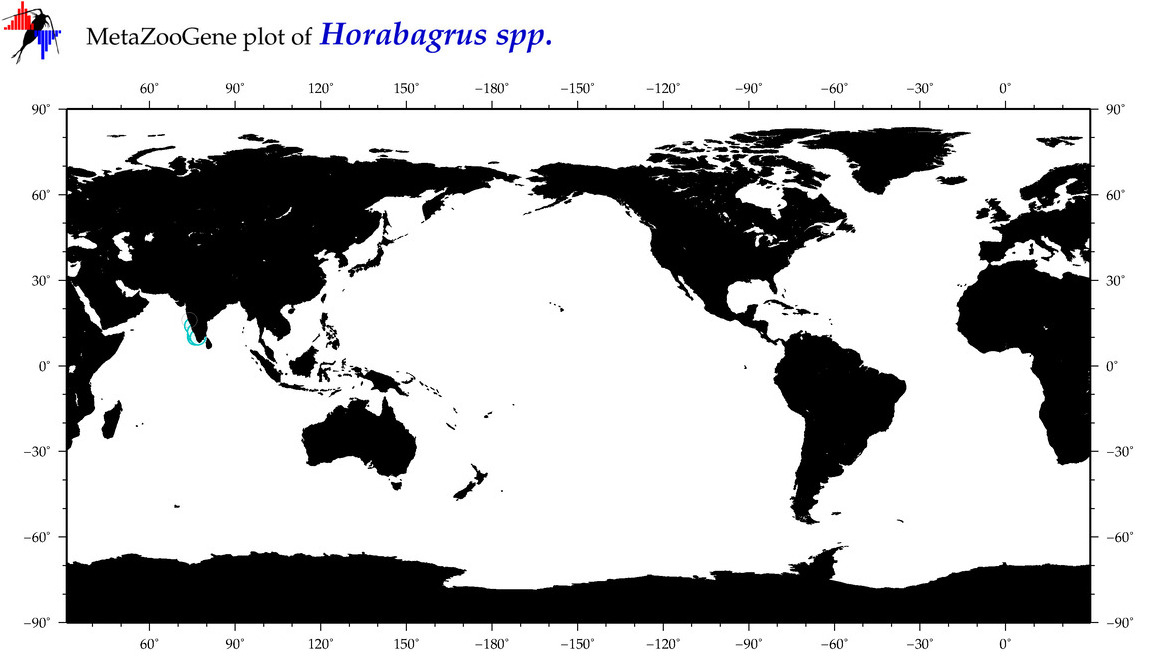 |
accepted
T5006815
R:0:1:1:0 |
| ⋄ Ictaluridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 57 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 57 |
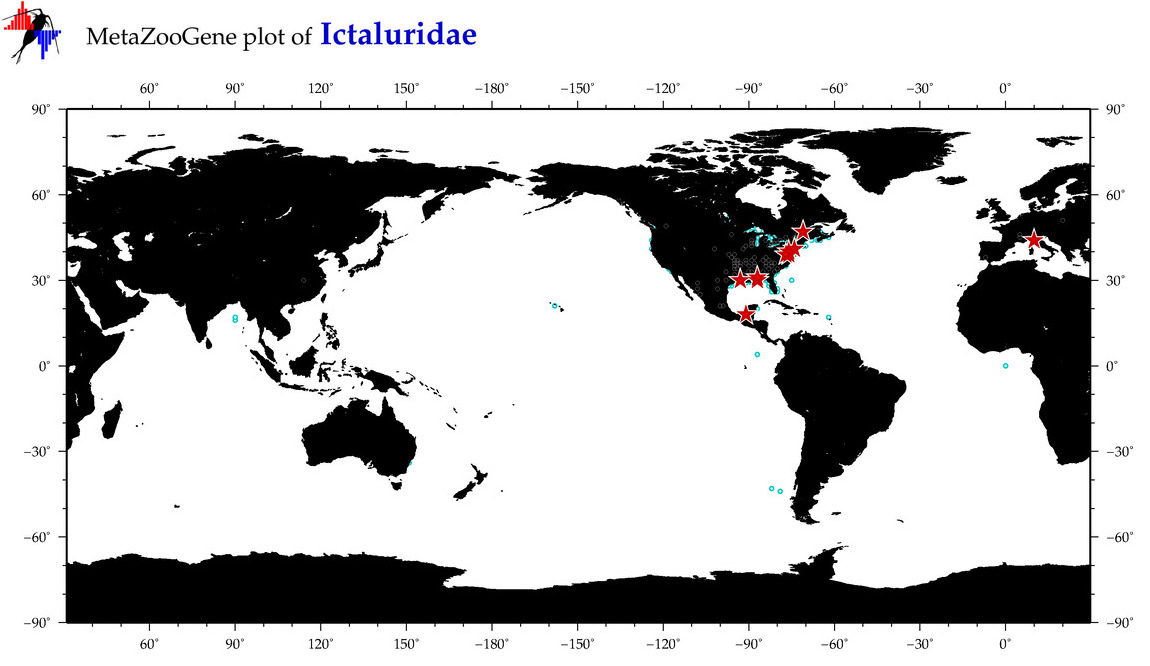 |
accepted
T5005955
R:0:1:1:0 |
| ⋄ ⋄ Ameiurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 17 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 17 |
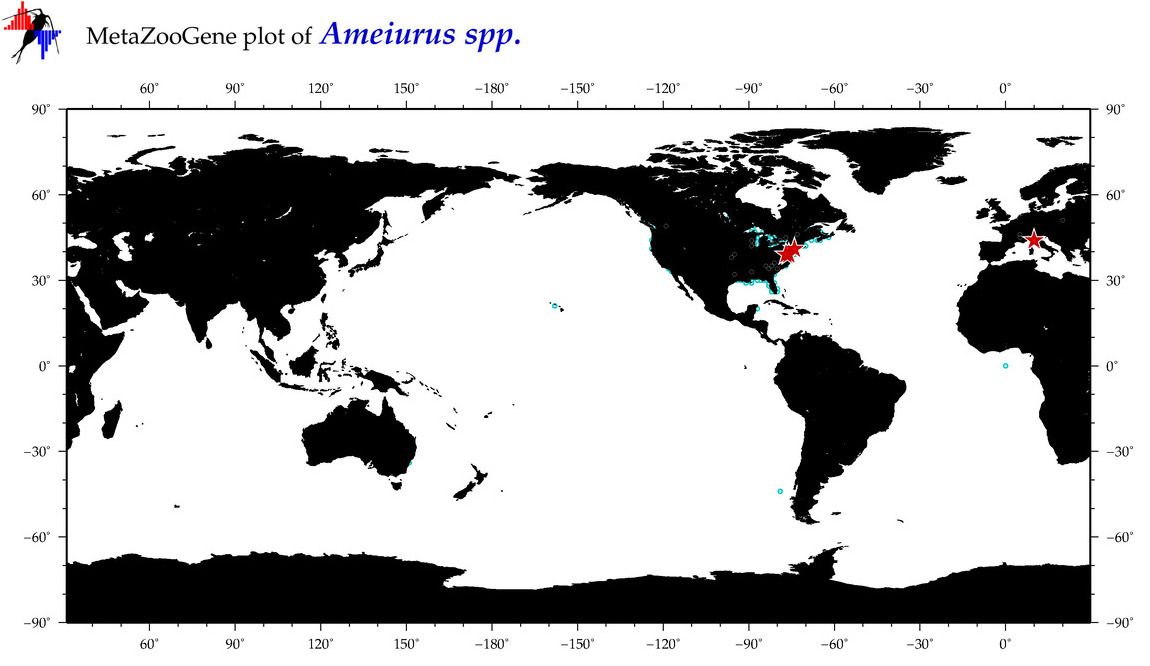 |
accepted
T5006259
R:1:1:1:0 |
| ⋄ ⋄ Ictalurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 40 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 40 |
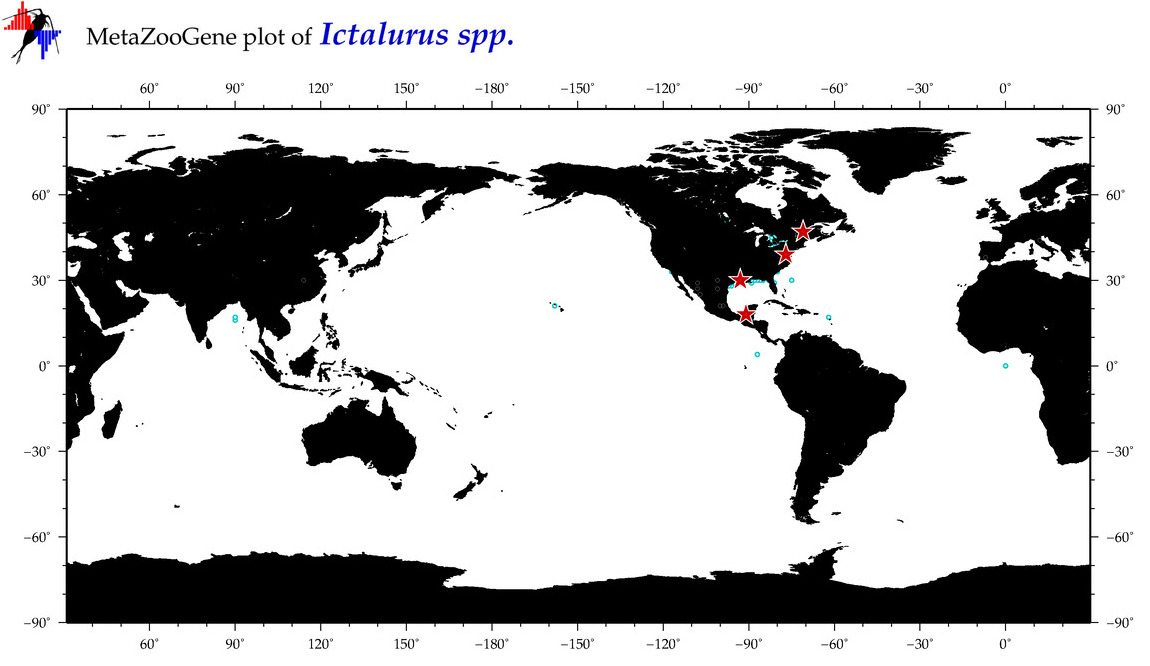 |
accepted
T5006261
R:0:1:1:0 |
| ⋄ Loricariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 5
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 23 |
Total Species: 5
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 23 |
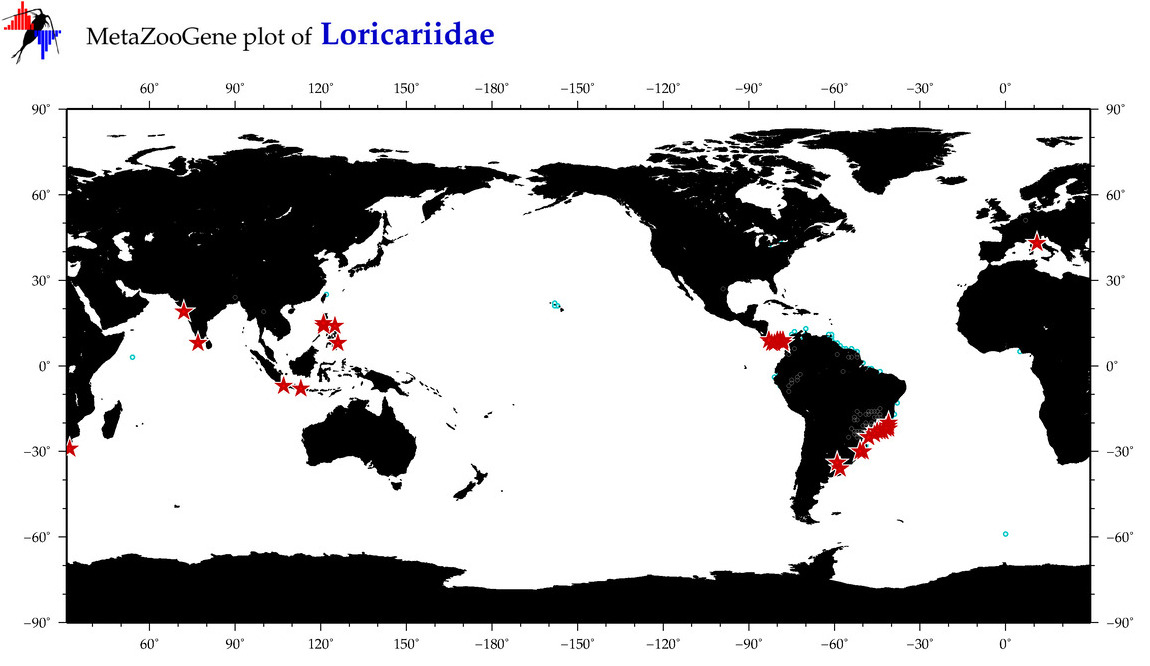 |
accepted
T5005999
R:1:1:1:0 |
| ⋄ ⋄ Hypoptopomatinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 5 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 5 |
 |
accepted
T5006144
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Hisonotus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 5 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 5 |
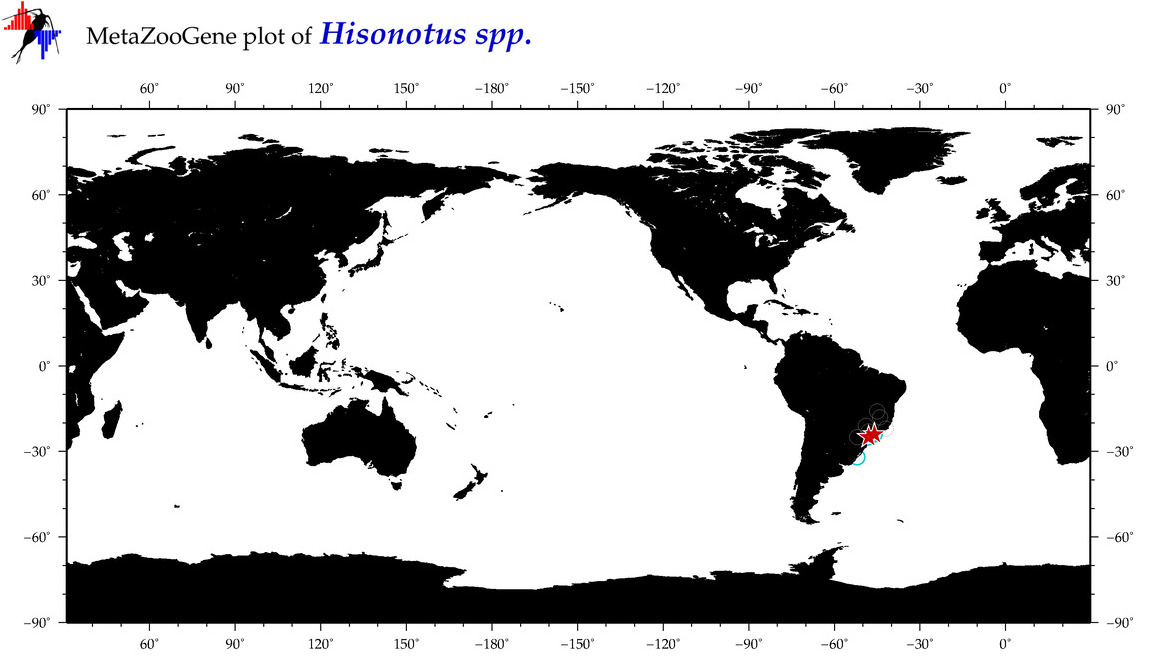 |
accepted
T5006807
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Nannoptopoma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5012850
R:1:0:1:0 |
| ⋄ ⋄ Hypostominae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 16 |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 16 |
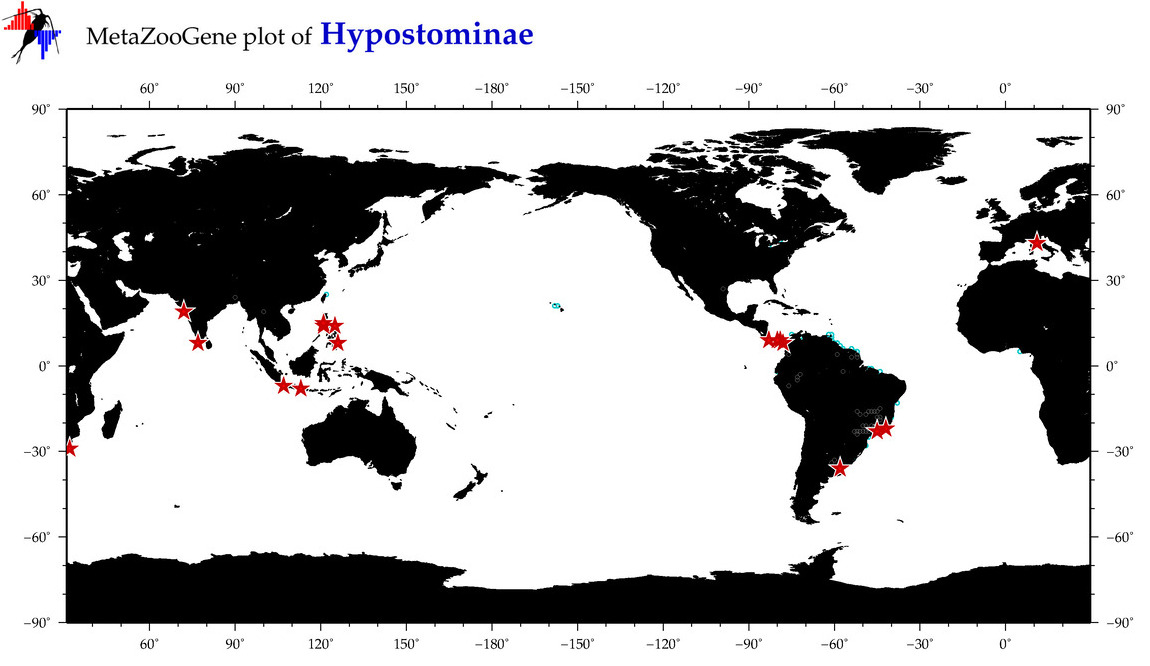 |
accepted
T5006142
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Cryptancistrus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5007538
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Hypostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 16 |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 16 |
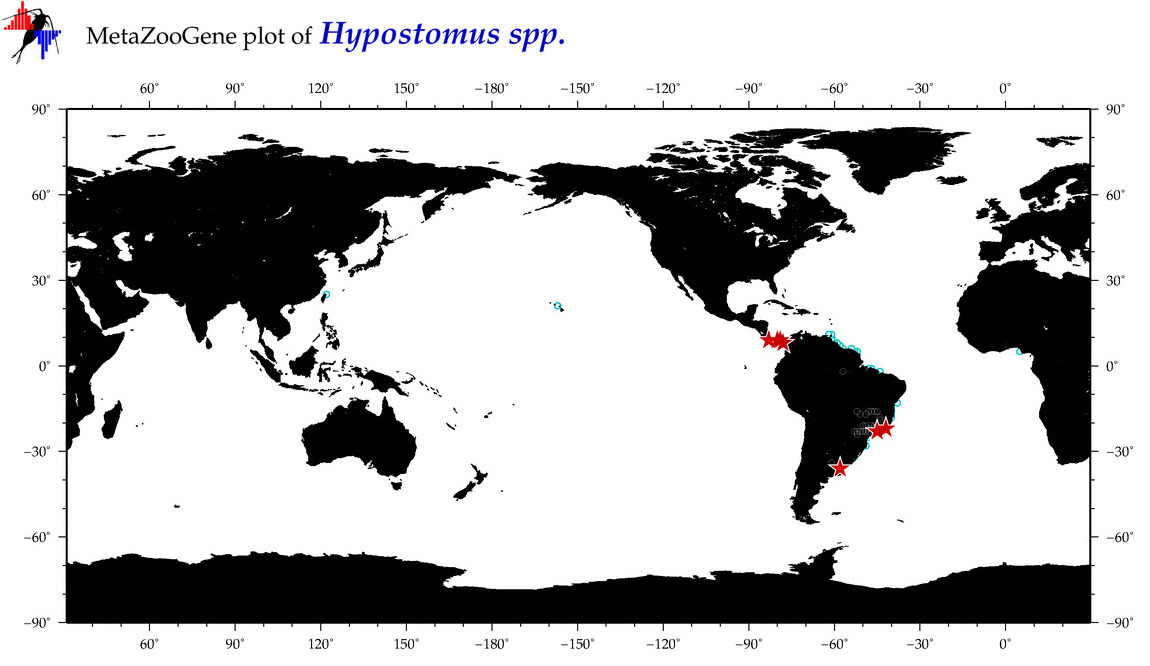 |
accepted
T5006836
R:0:1:1:0 |
| ⋄ ⋄ Loricariinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
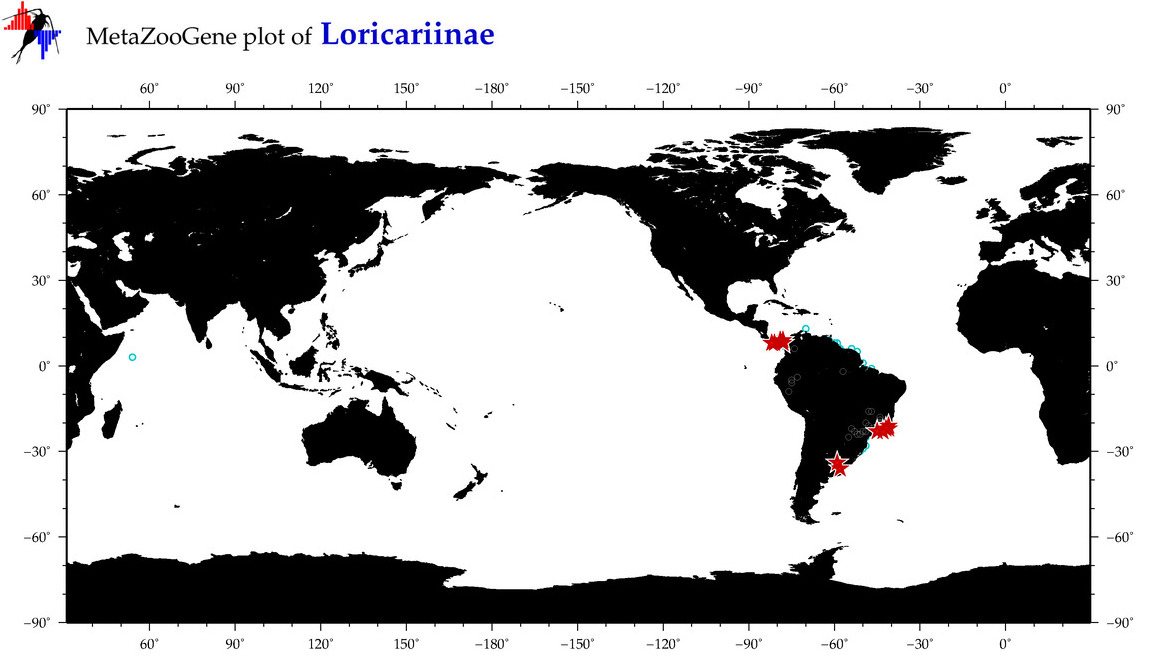 |
accepted
T5006139
R:1:0:1:0 |
| ⋄ ⋄ Nannoplecostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
 |
accepted
T5007514
R:1:0:1:0 |
| ⋄ ⋄ ⋄ Nannoplecostomus eleonorae |
Species
(87) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 2
18S = 0
28S = 0
|
 |
accepted
T5010591
R:1:0:1:0 |
| ⋄ ⋄ Neoplecostominae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
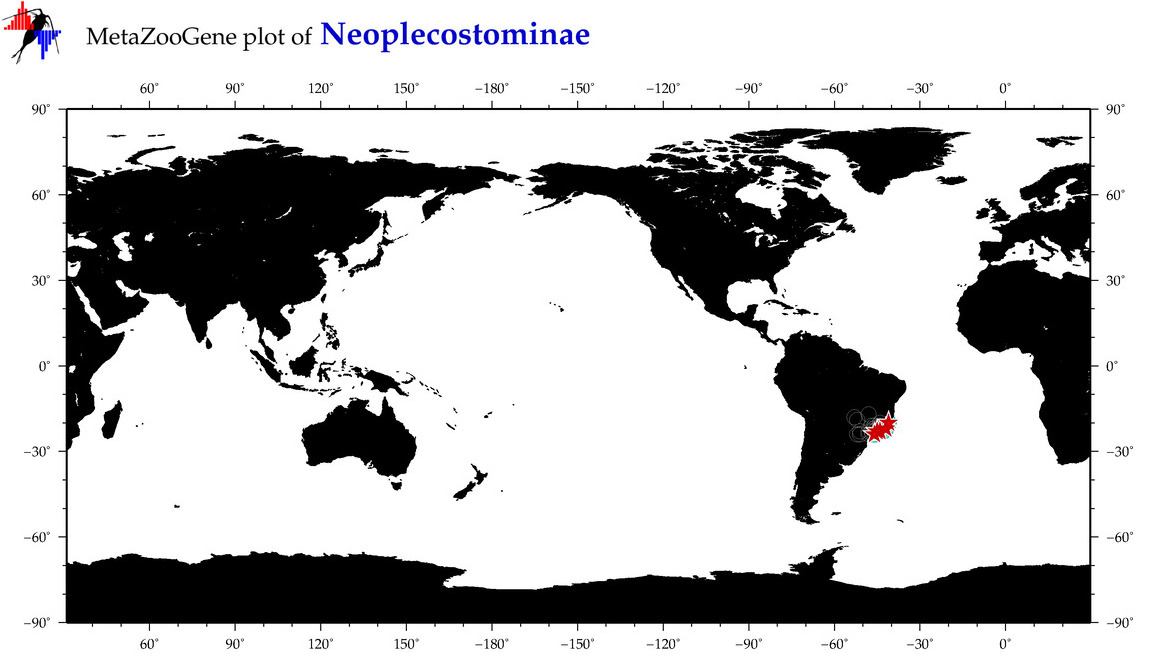 |
accepted
T5006140
R:0:1:1:0 |
| ⋄ Mochokidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
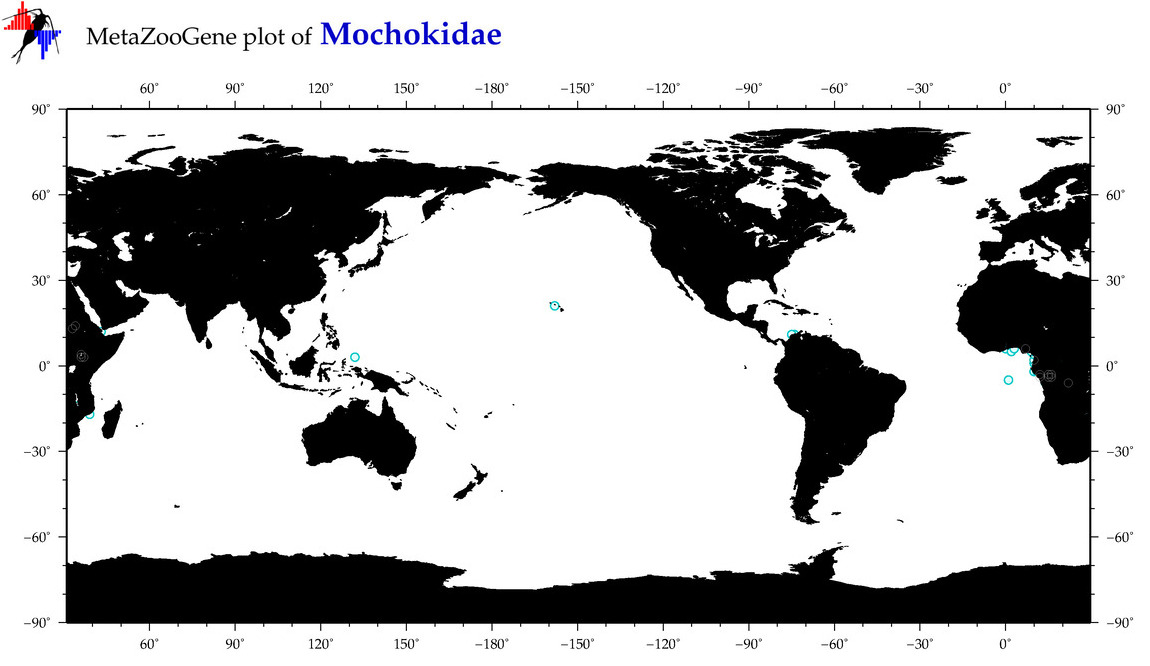 |
accepted
T5005994
R:0:1:1:0 |
| ⋄ ⋄ Chiloglanidinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
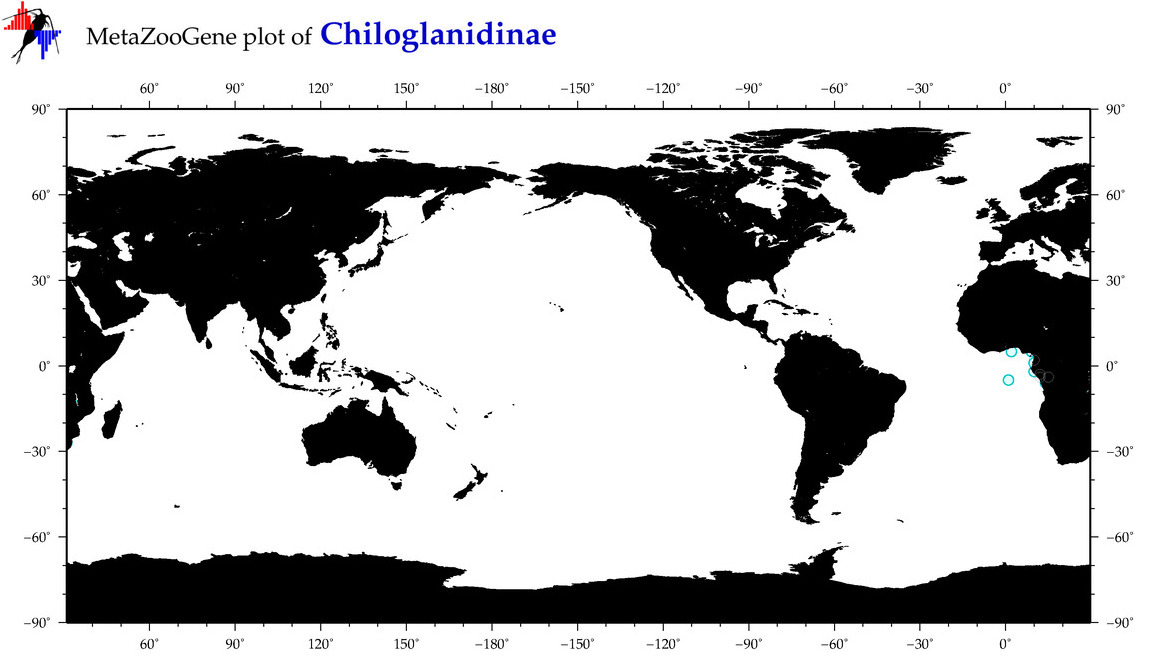 |
accepted
T5006185
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Chiloglanis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
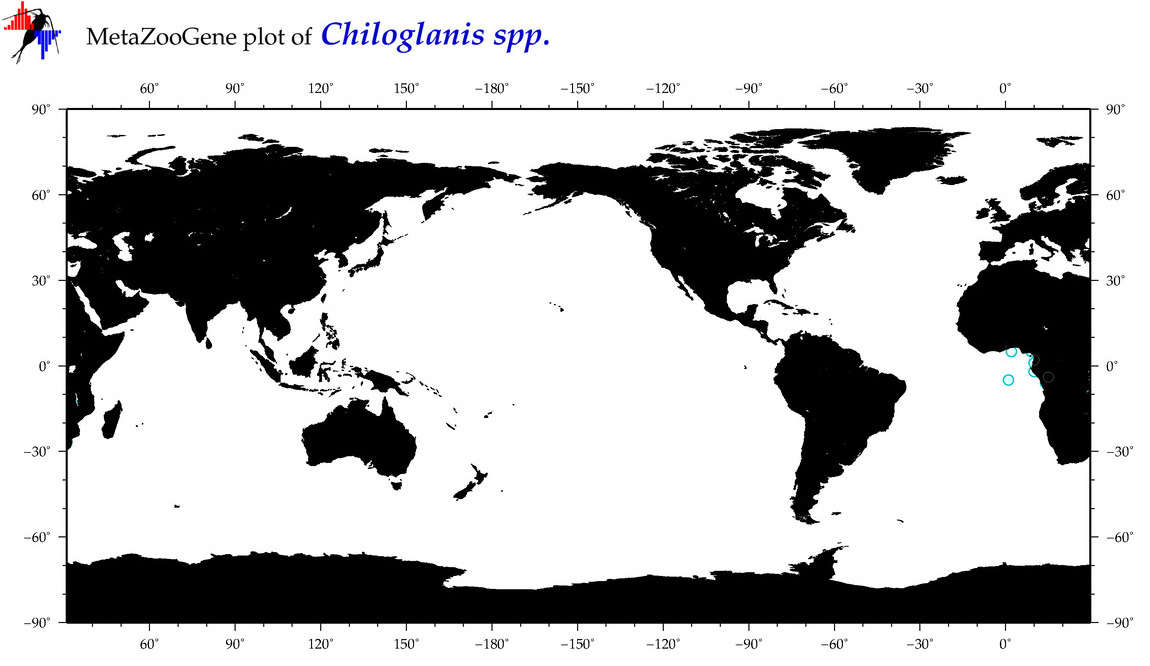 |
accepted
T5006608
R:0:1:1:0 |
| ⋄ Pangasiidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 6
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 62 |
Total Species: 6
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 62 |
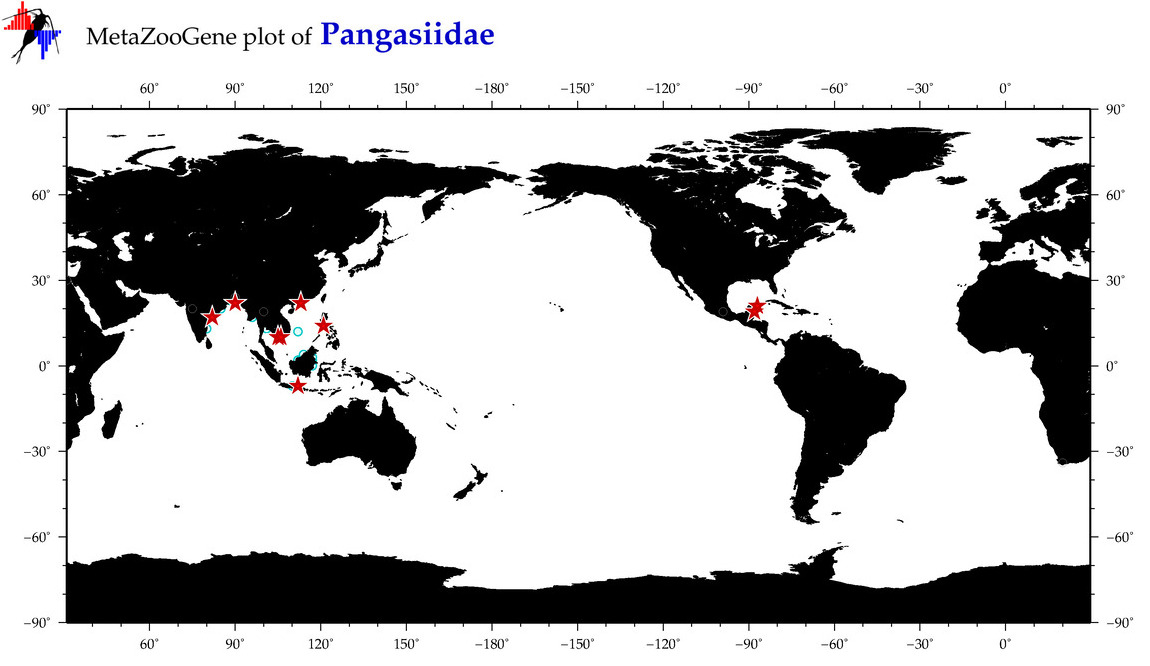 |
accepted
T5006019
R:1:1:1:0 |
| ⋄ ⋄ Pangasius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 5
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 61 |
Total Species: 5
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 61 |
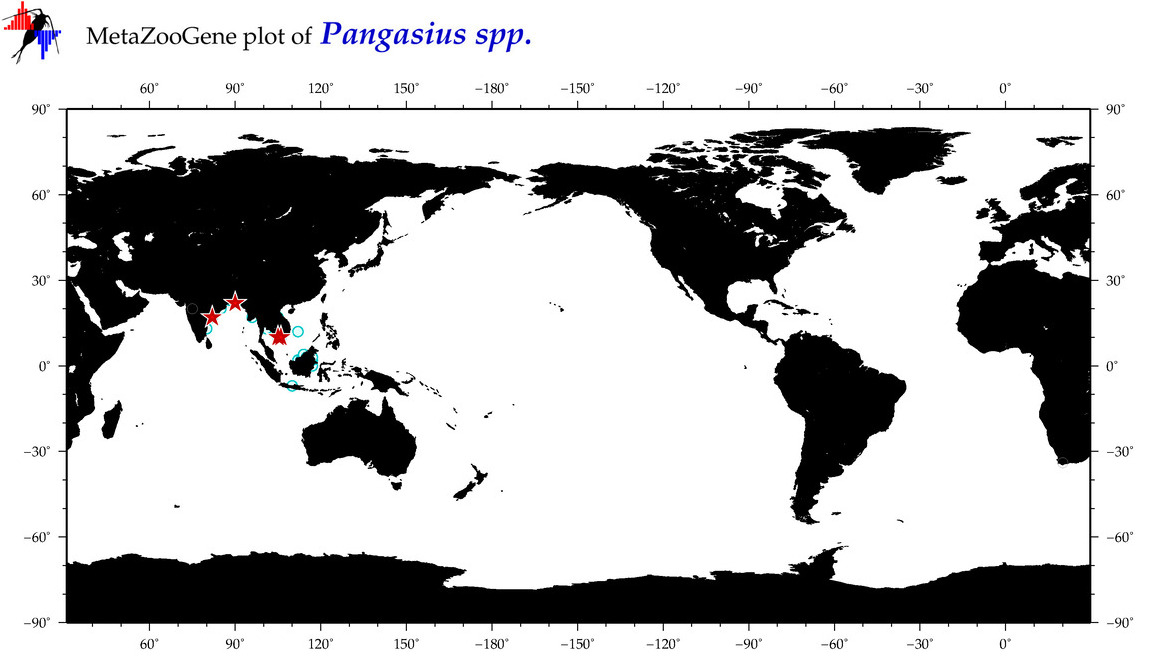 |
accepted
T5007065
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Pangasius krempfi |
Species
(88) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 3
16S = 3
18S = 0
28S = 0
|
COI = 6
12S = 3
16S = 3
18S = 0
28S = 0
|
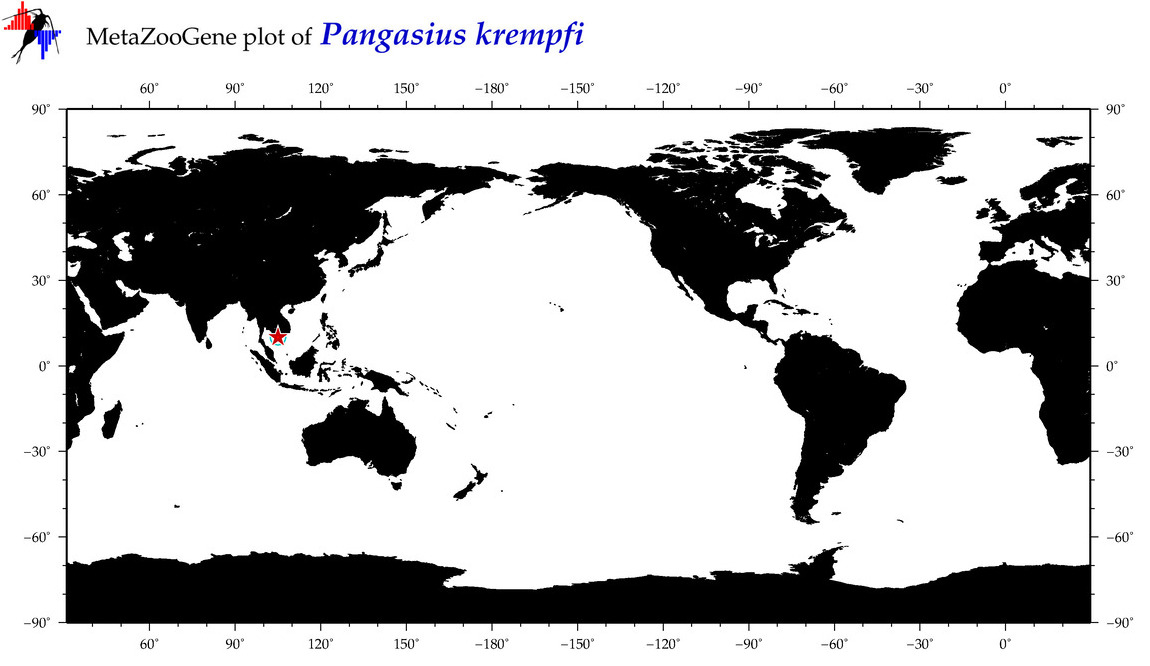 |
accepted
T5011082
R:1:1:1:0 |
| ⋄ ⋄ Pseudolais |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
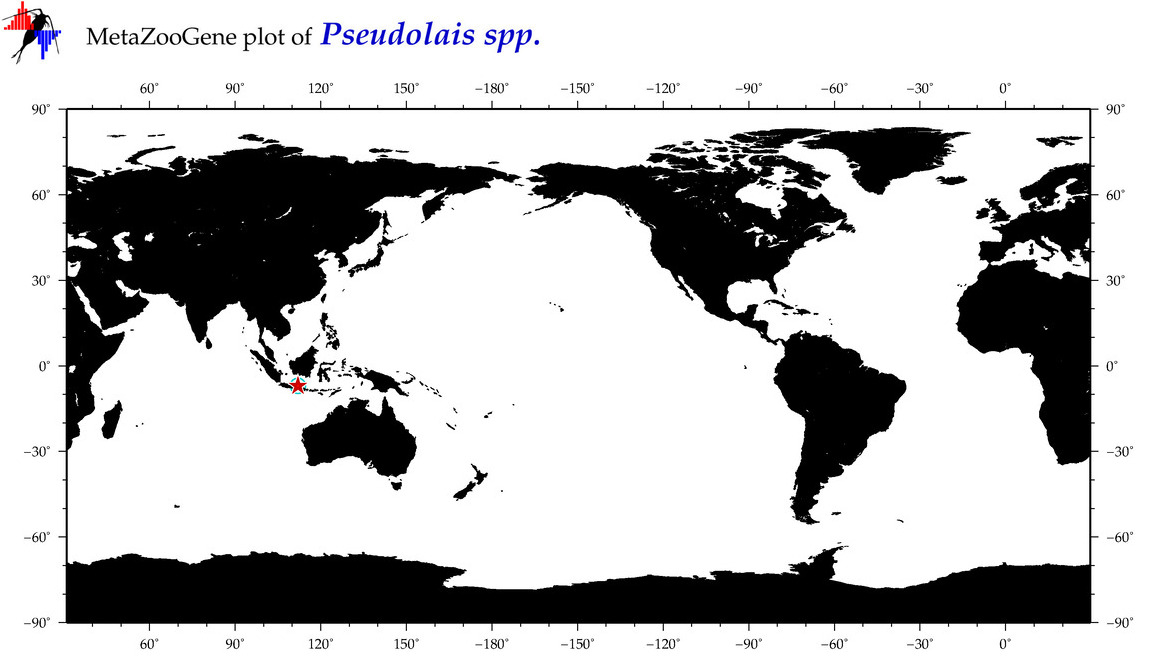 |
accepted
T5007446
R:0:1:1:0 |
| ⋄ Pimelodidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 6
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 225 |
Total Species: 6
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 225 |
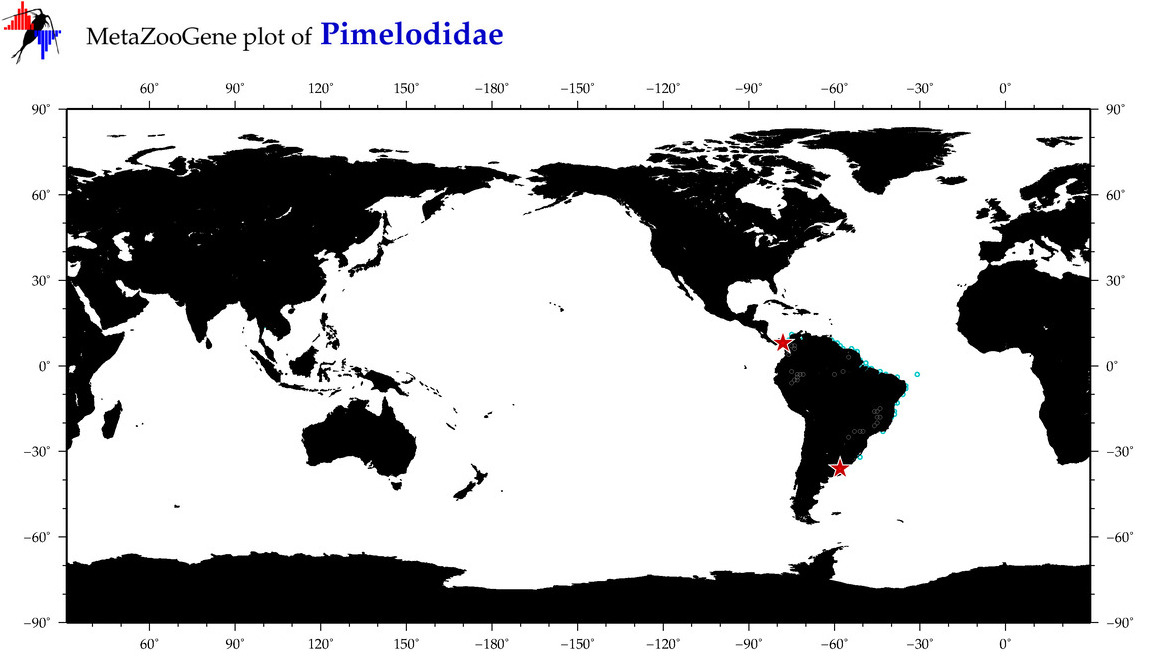 |
accepted
T5005980
R:0:1:1:0 |
| ⋄ ⋄ Brachyplatystoma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 62 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 62 |
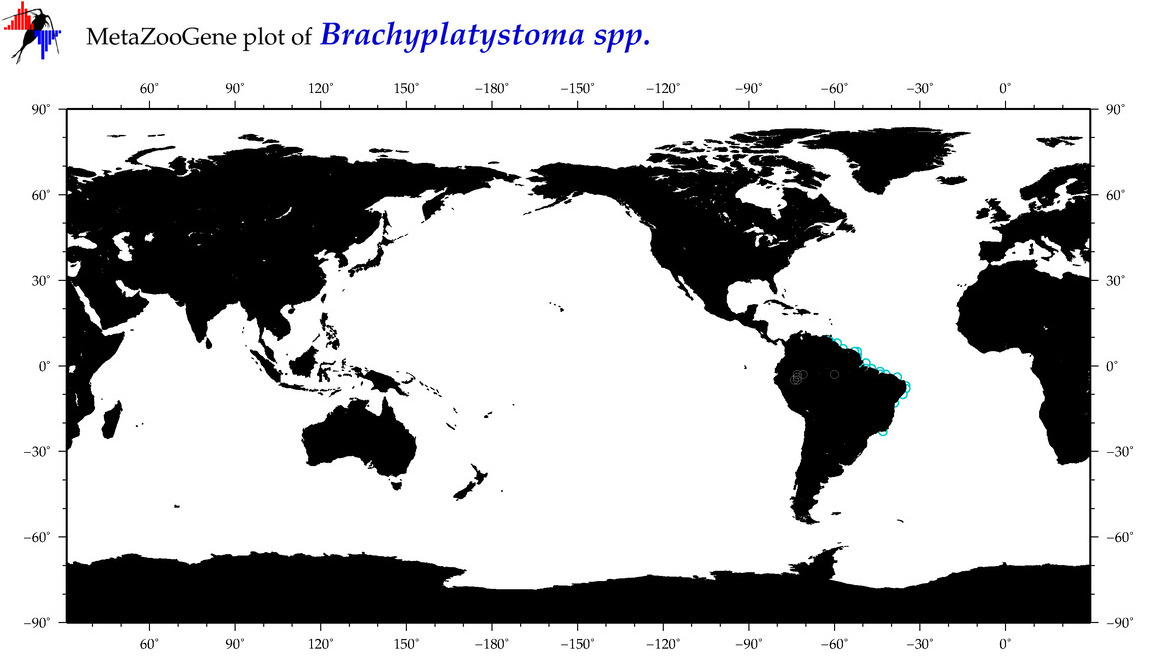 |
accepted
T5006544
R:0:1:1:0 |
| ⋄ ⋄ Pimelodus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 54 |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 54 |
 |
accepted
T5007140
R:0:1:1:0 |
| ⋄ ⋄ Pseudoplatystoma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 109 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 109 |
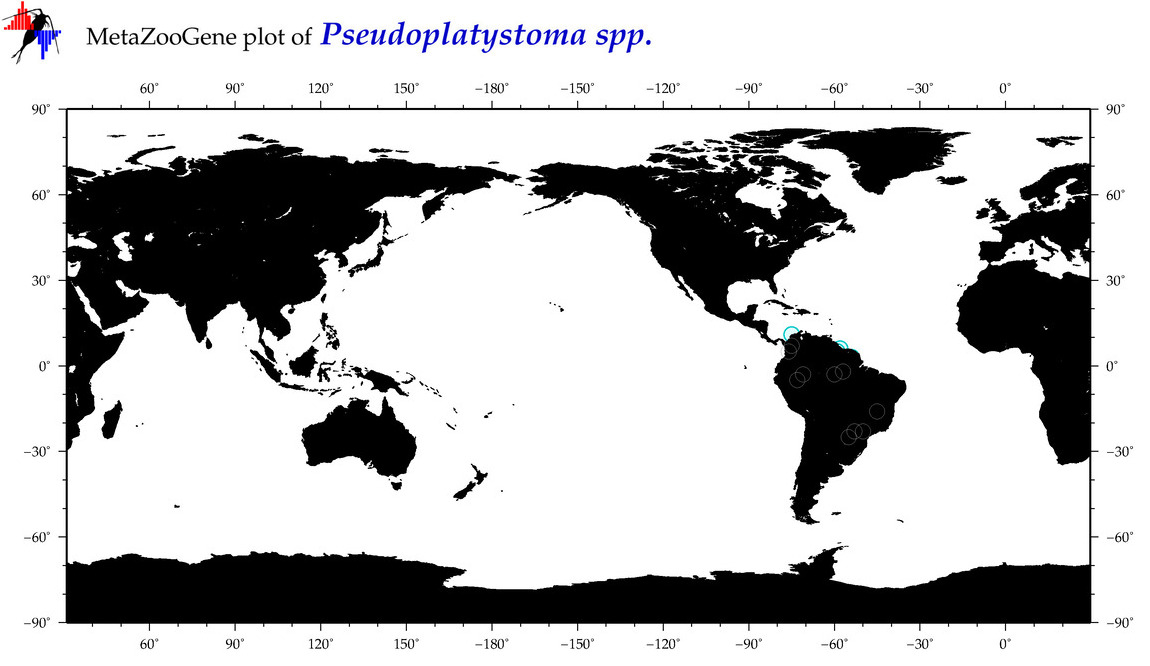 |
accepted
T5007212
R:0:1:1:0 |
| ⋄ Plotosidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 13
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 79 |
Total Species: 13
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 79 |
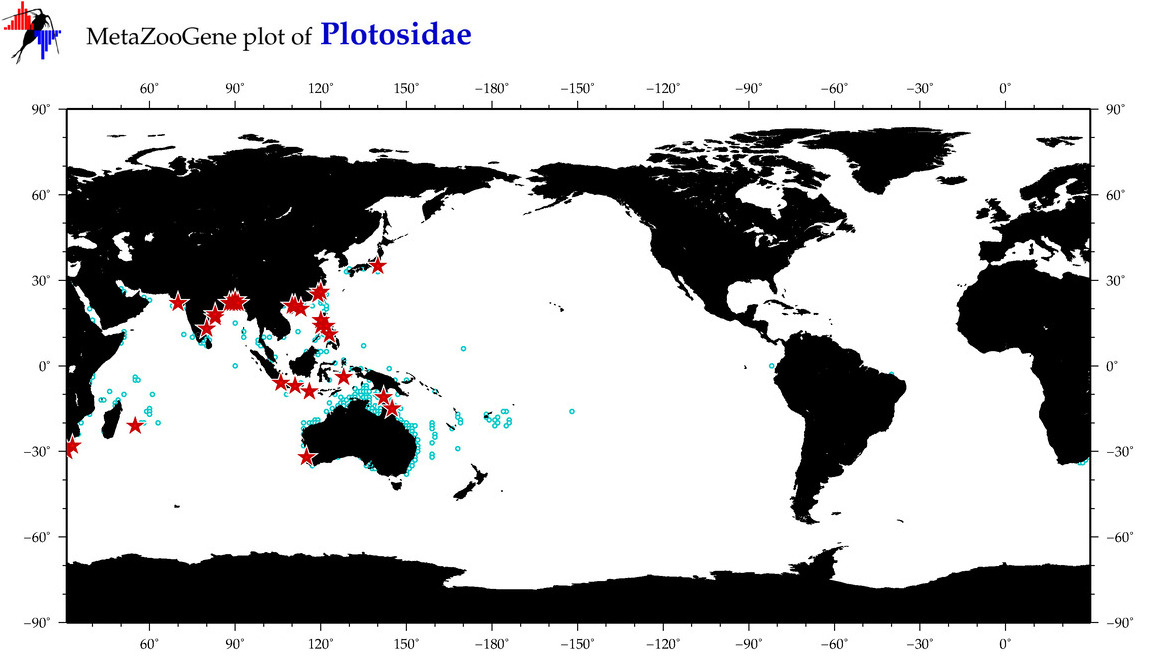 |
accepted
T5005978
R:1:1:1:0 |
| ⋄ ⋄ Cnidoglanis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
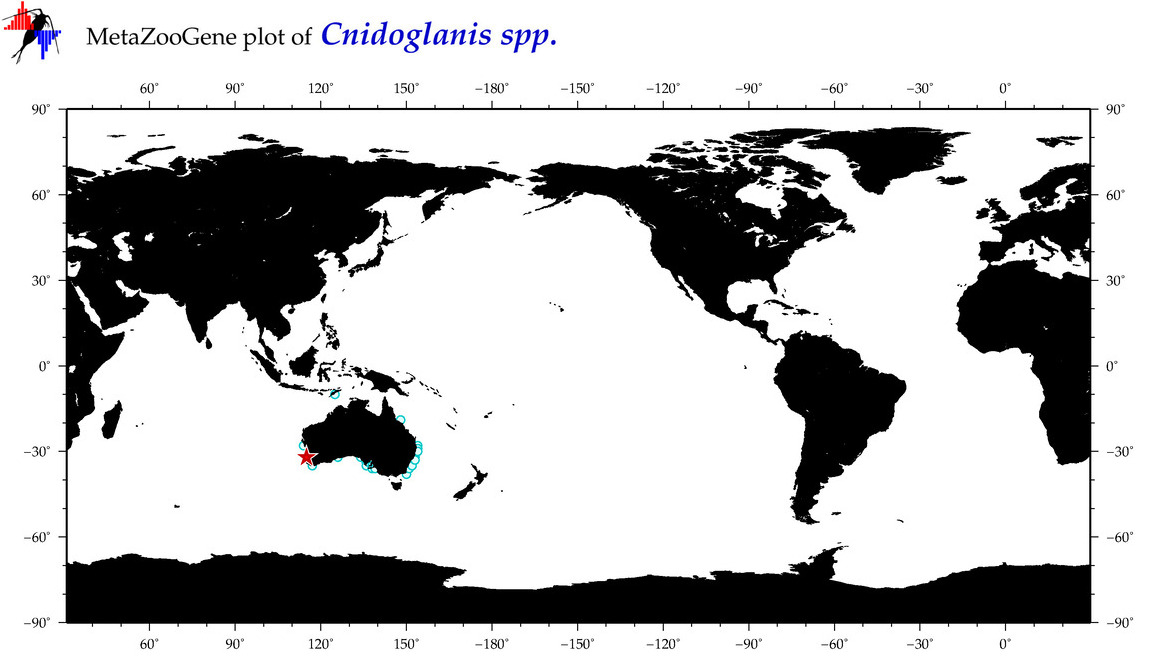 |
accepted
T5006624
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Cnidoglanis macrocephalus |
Species
(89) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 1
18S = 2
28S = 0
|
COI = 2
12S = 1
16S = 1
18S = 2
28S = 0
|
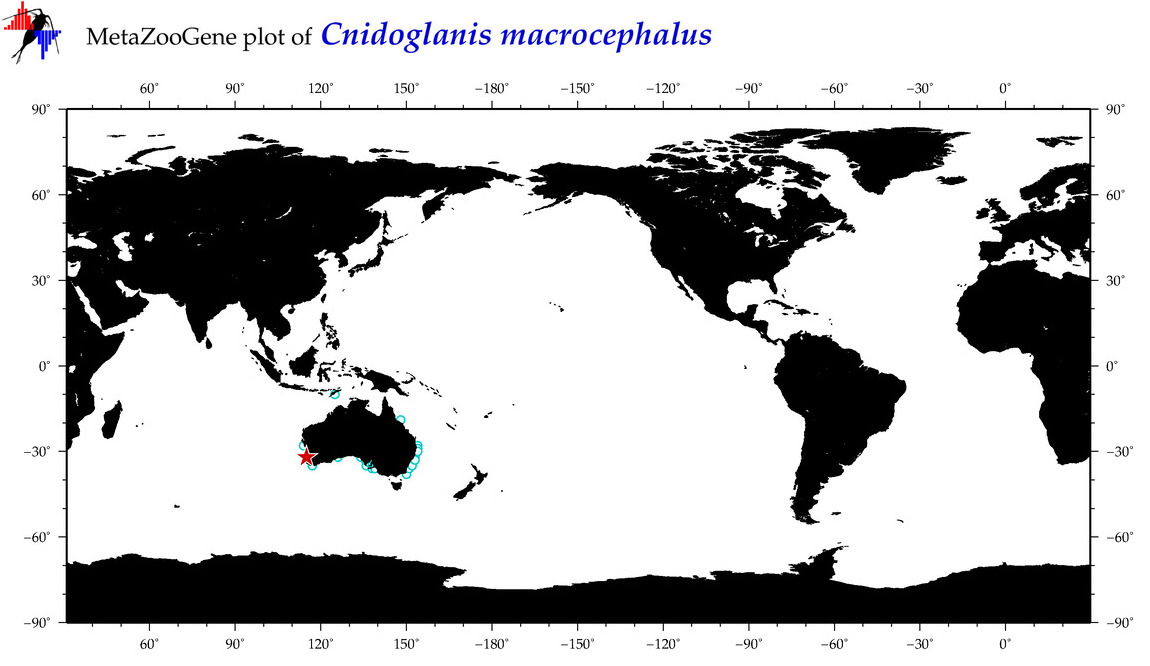 |
accepted
T5008564
R:1:1:0:0 |
| ⋄ ⋄ Euristhmus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 5
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
Total Species: 5
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
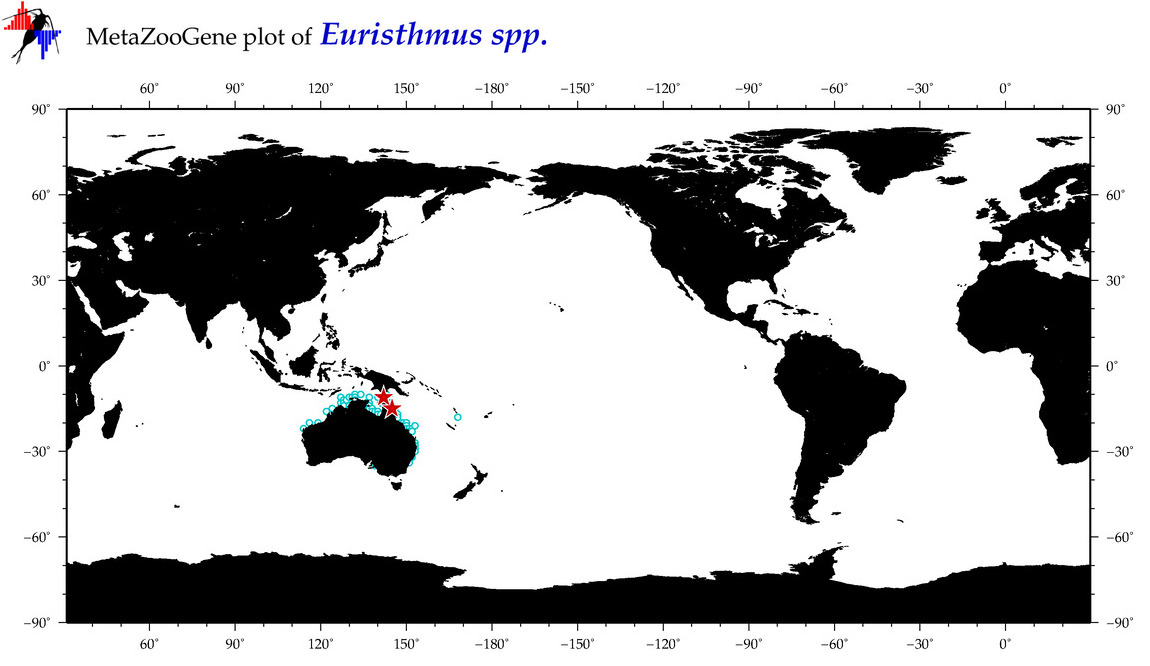 |
accepted
T5006719
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Euristhmus lepturus |
Species
(90) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
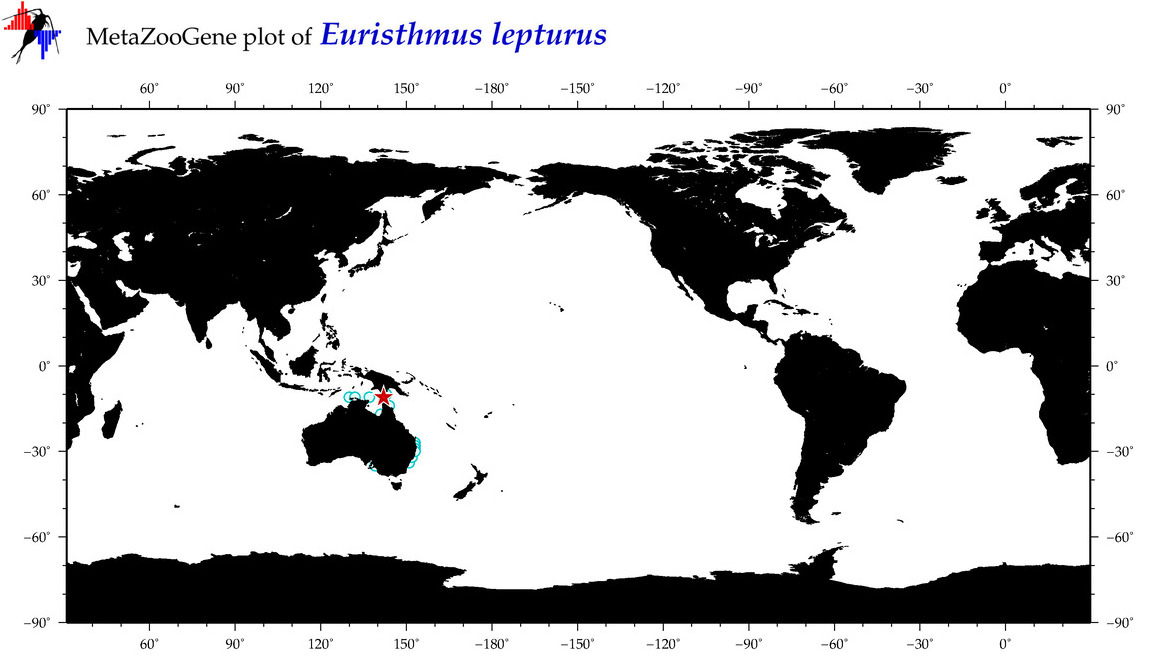 |
accepted
T5009215
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Euristhmus microceps |
Species
(91) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019700
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Euristhmus microphthalmus |
Species
(92) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028023
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Euristhmus nudiceps |
Species
(93) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
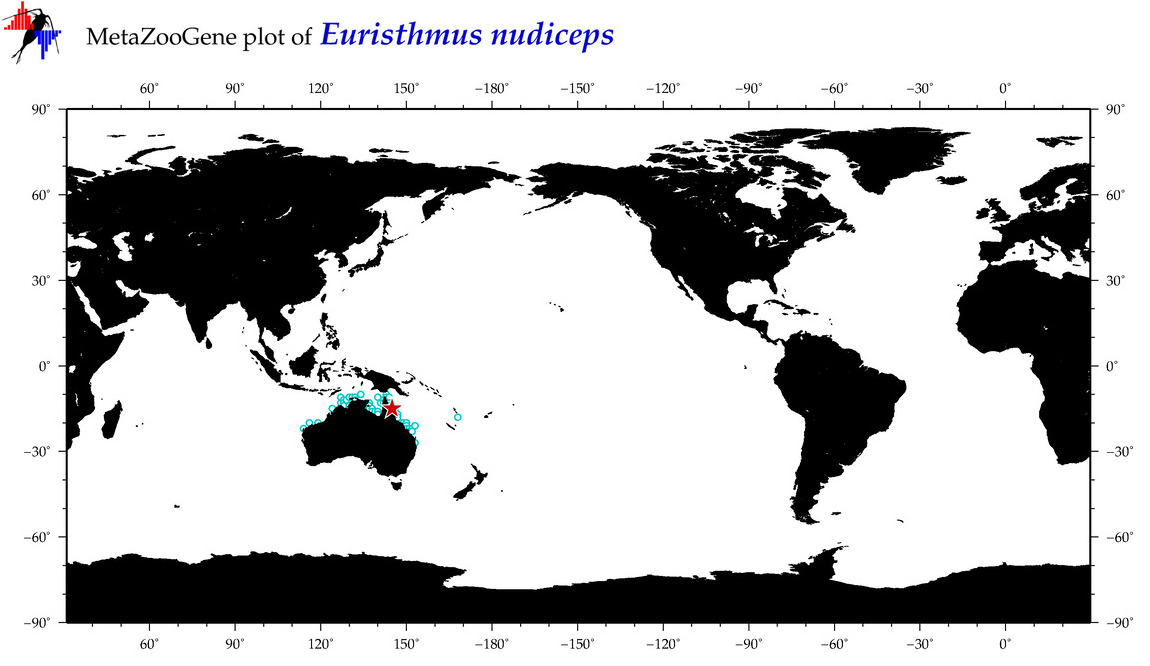 |
accepted
T5009216
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Euristhmus sandrae |
Species
(94) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028024
R:1:0:0:0 |
| ⋄ ⋄ Paraplotosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
Total Species: 3
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
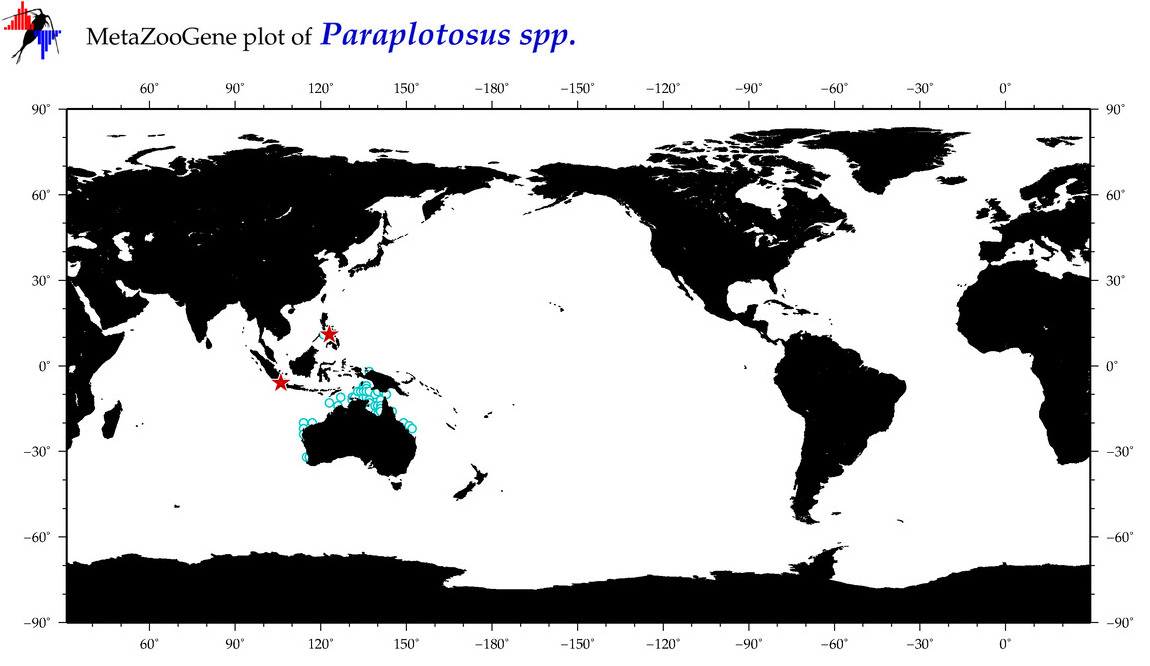 |
accepted
T5007088
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Paraplotosus albilabris |
Species
(95) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 9
12S = 0
16S = 0
18S = 0
28S = 0
|
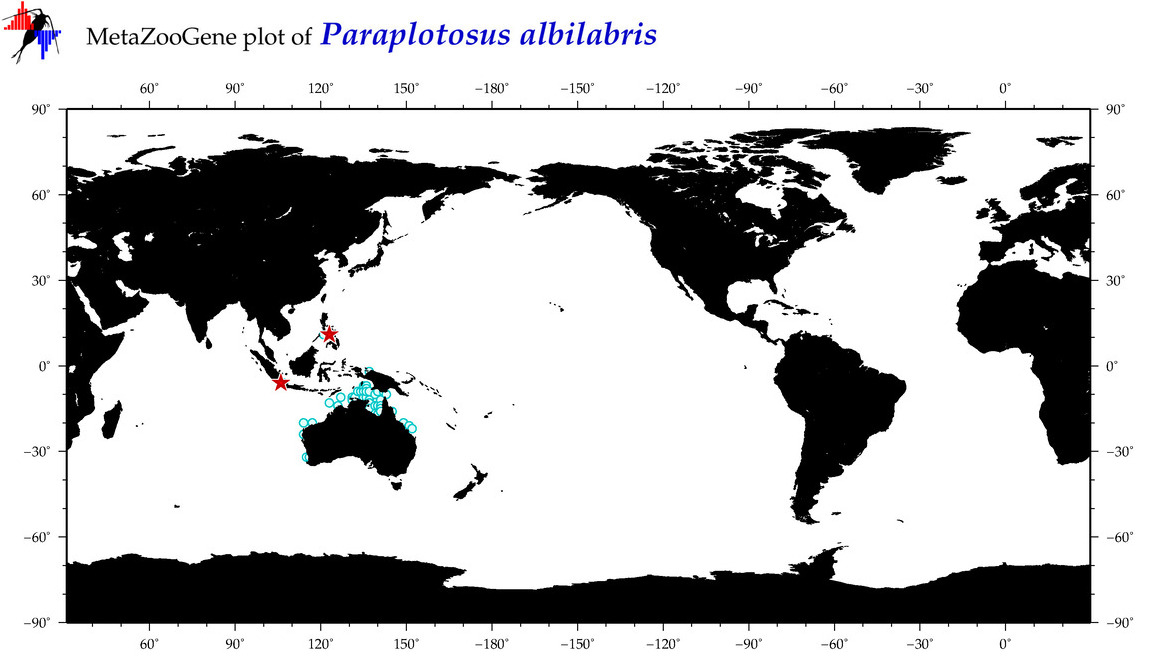 |
accepted
T5011154
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Paraplotosus butleri |
Species
(96) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5023620
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Paraplotosus muelleri |
Species
(97) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028444
R:1:0:0:0 |
| ⋄ ⋄ Plotosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 65 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 65 |
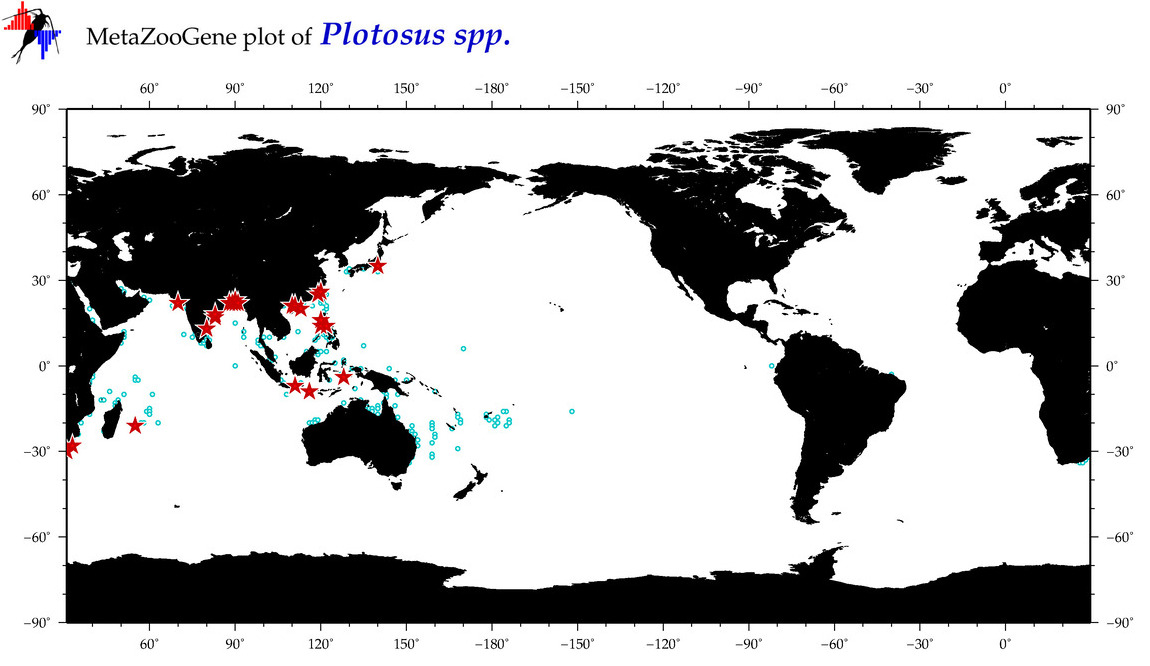 |
accepted
T5006354
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Plotosus canius |
Species
(98) |
COI
12S
16S
18S
28S
|
COI = 60
12S = 2
16S = 0
18S = 0
28S = 0
|
COI = 60
12S = 2
16S = 0
18S = 0
28S = 0
|
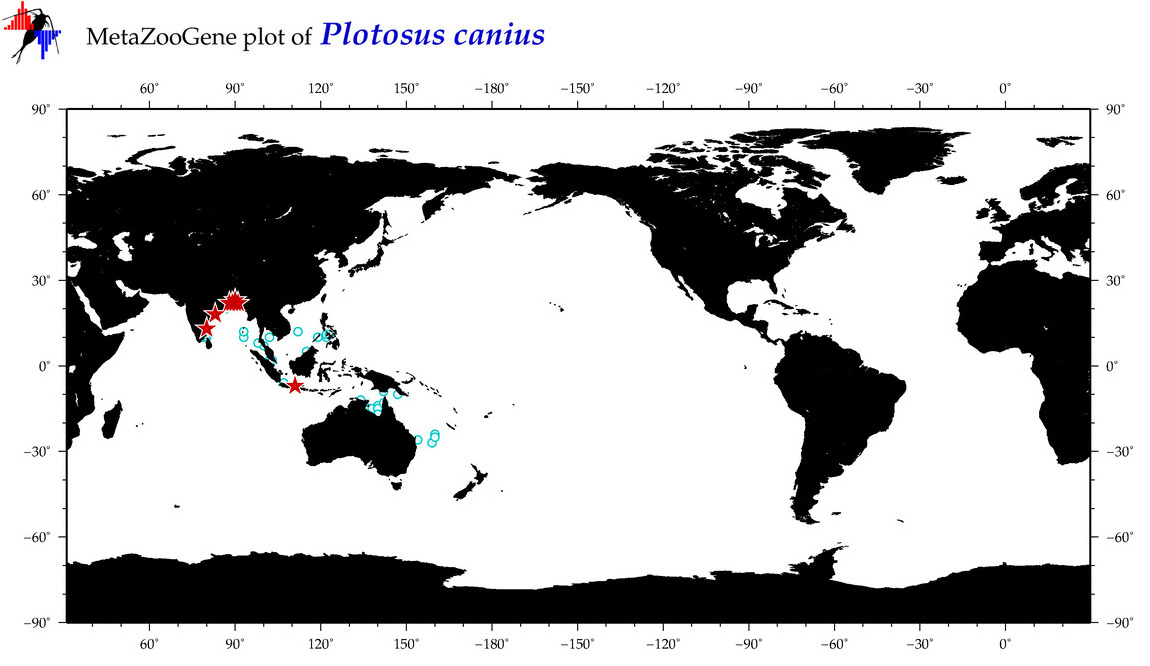 |
accepted
T5011476
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Plotosus japonicus |
Species
(99) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 13
16S = 4
18S = 0
28S = 0
|
COI = 2
12S = 13
16S = 4
18S = 0
28S = 0
|
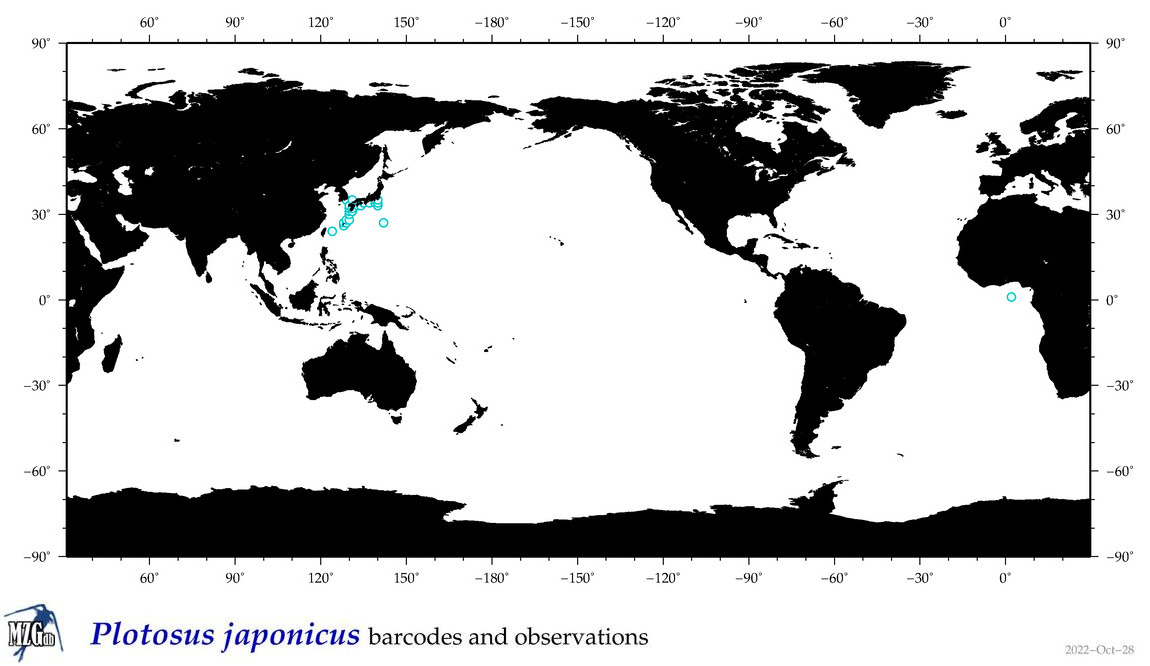 |
accepted
T5028499
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Plotosus limbatus |
Species
(100) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
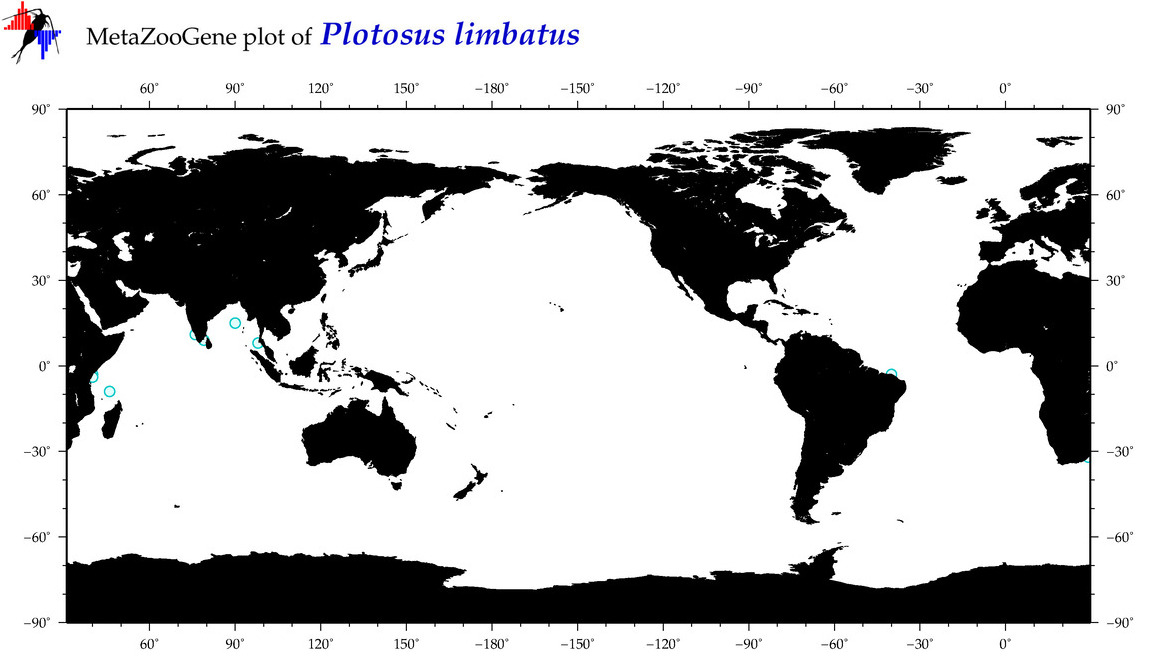 |
accepted
T5011477
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Plotosus nkunga |
Species
(101) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
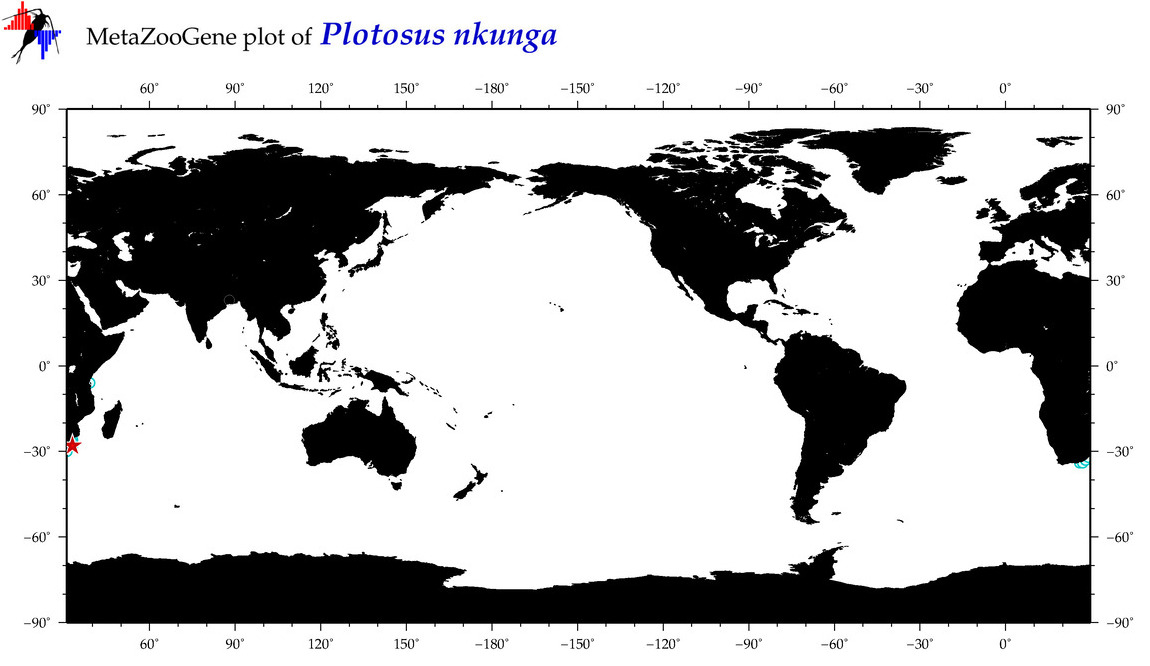 |
accepted
T5011478
R:1:1:1:0 |
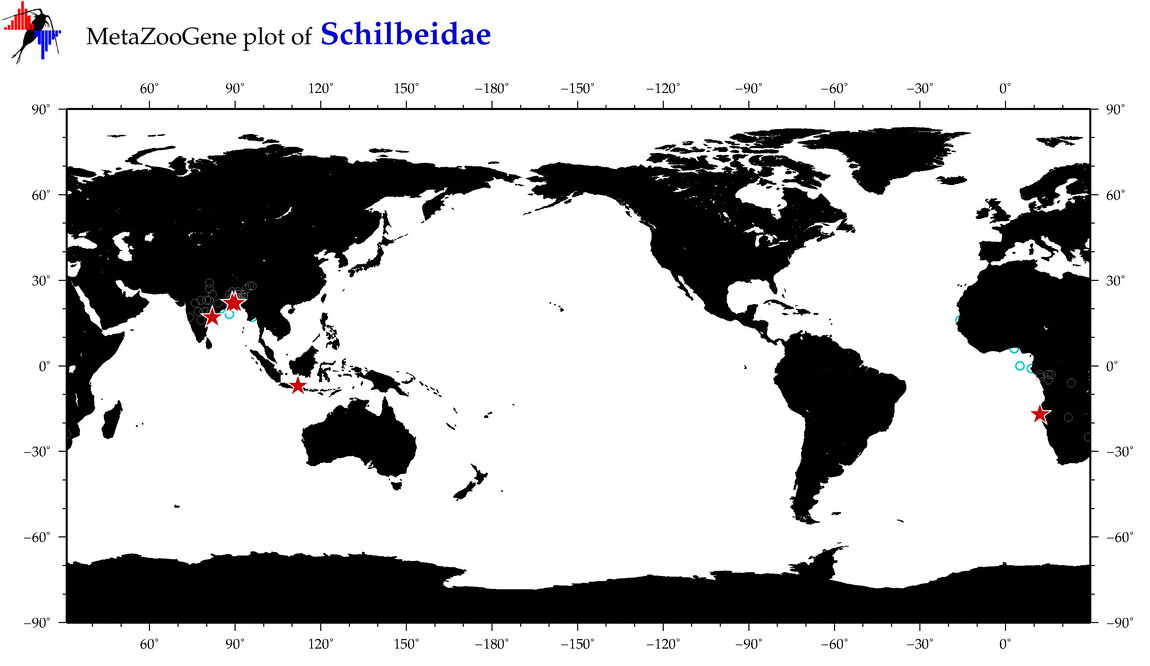
| ⋄ Schilbeidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 5
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 133 |
Total Species: 5
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 133 |
 |
accepted
T5006018
R:1:1:1:0 |
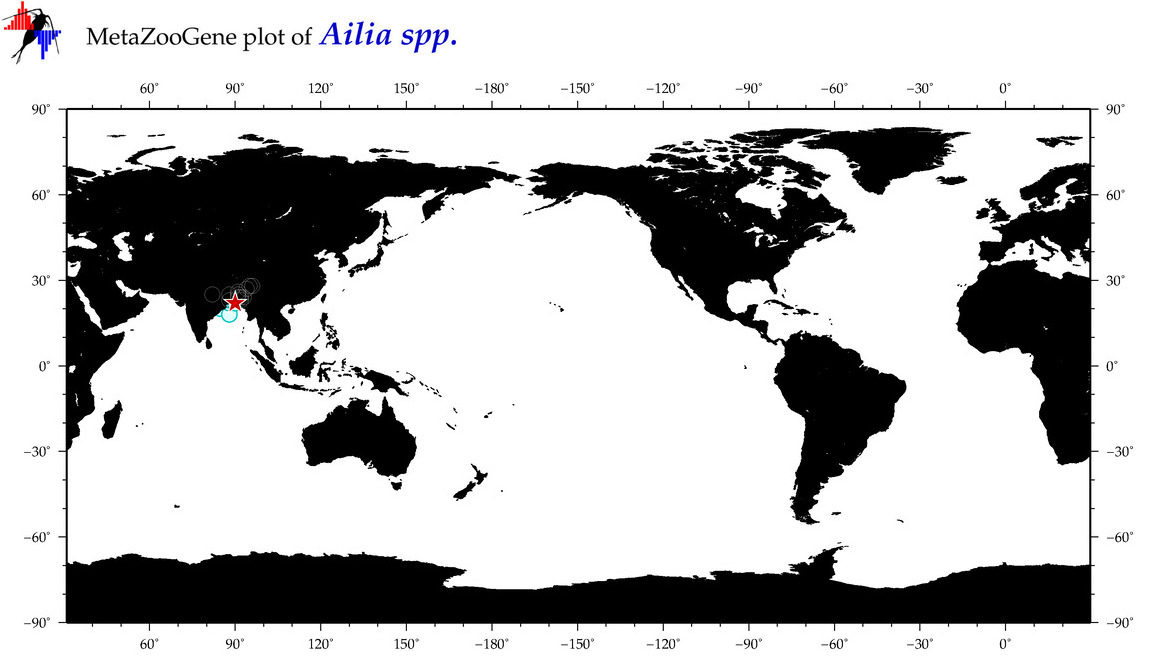
| ⋄ ⋄ Ailia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 54 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 54 |
 |
accepted
T5006435
R:0:1:1:0 |
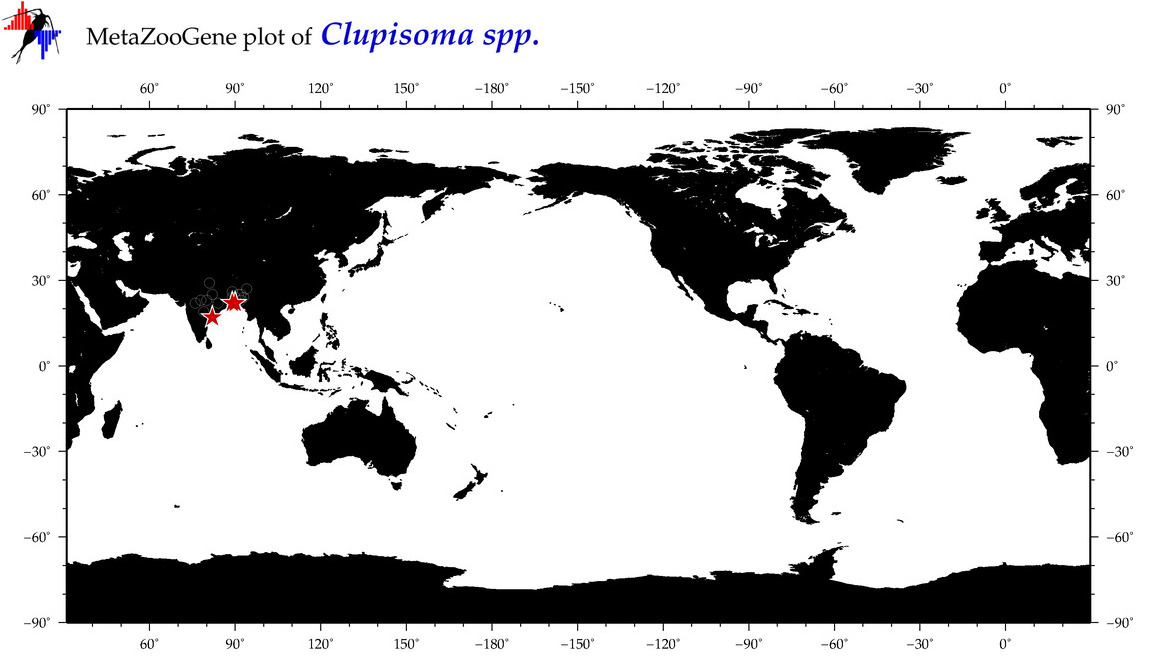
| ⋄ ⋄ Clupisoma |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 33 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 33 |
 |
accepted
T5006623
R:0:1:1:0 |
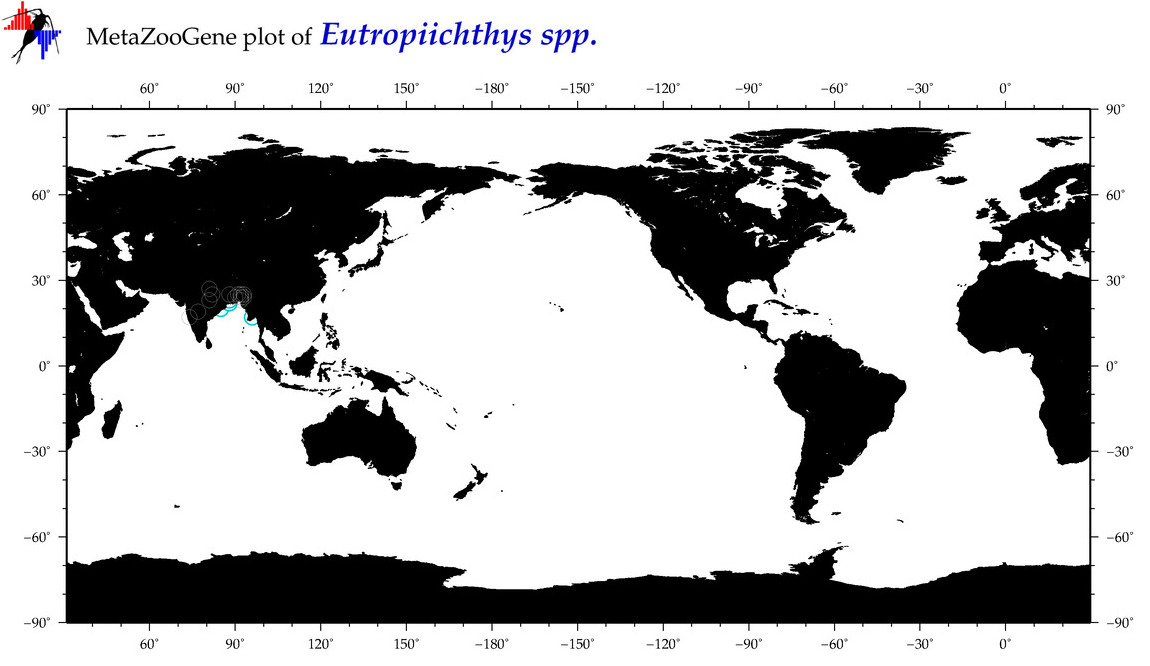
| ⋄ ⋄ Eutropiichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 35 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 35 |
 |
accepted
T5006721
R:0:1:1:0 |
| ⋄ ⋄ Schilbe |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
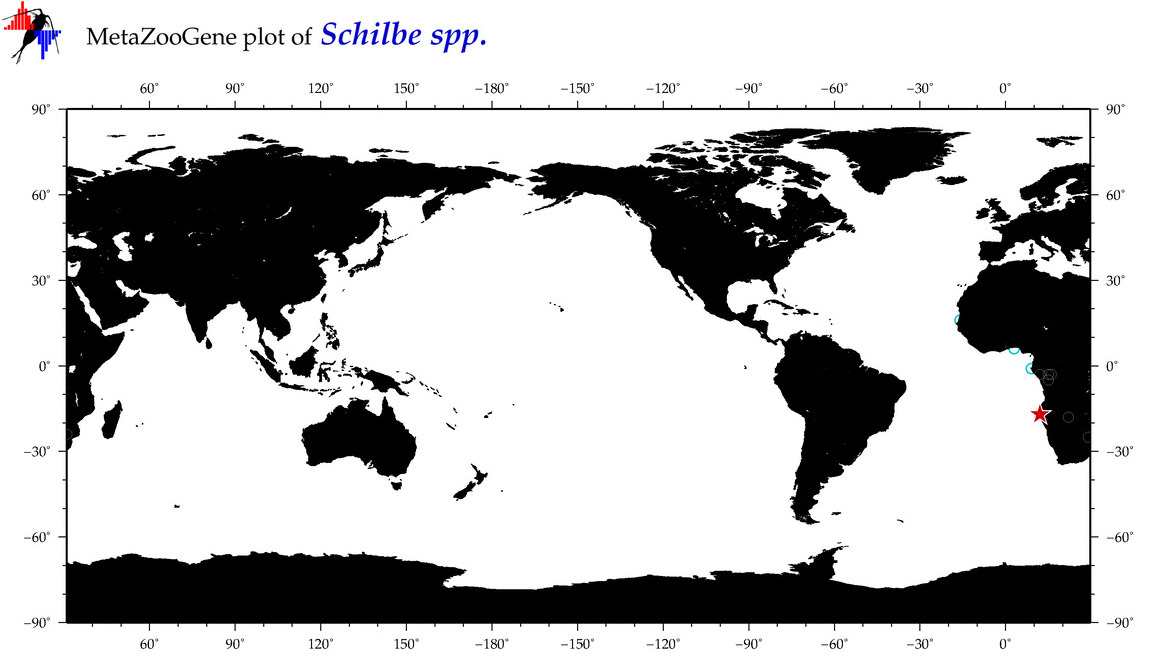 |
accepted
T5007295
R:0:1:1:0 |
| ⋄ ⋄ Silonia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 11 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 11 |
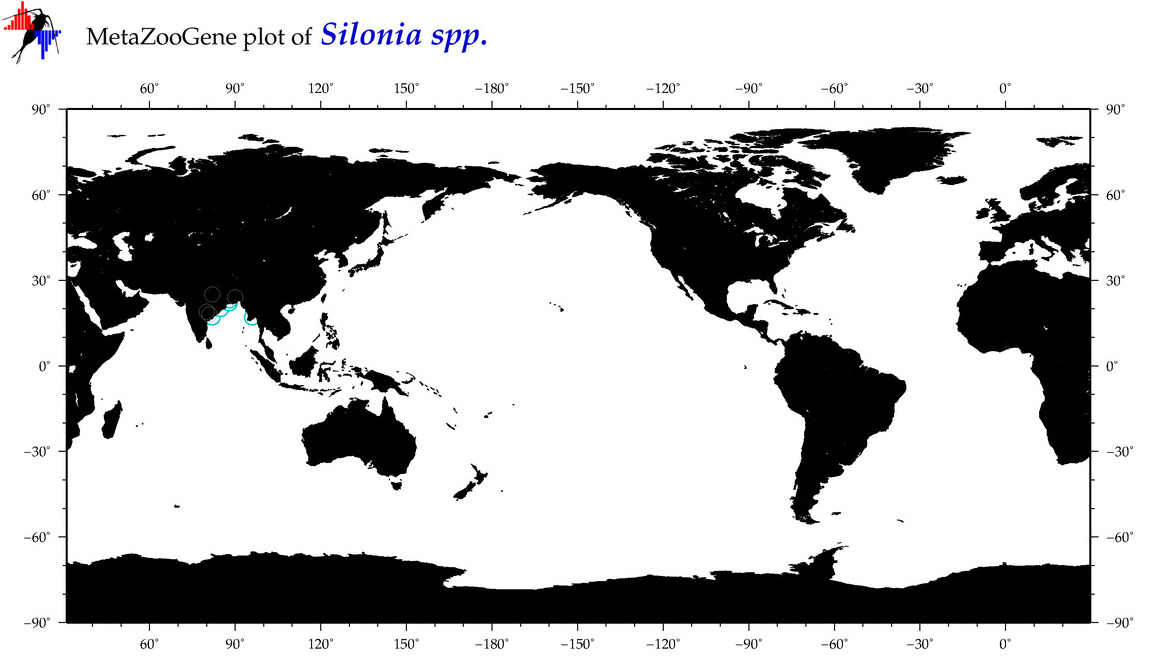 |
accepted
T5007319
R:0:1:1:0 |
| ⋄ Siluridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 6
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 369 |
Total Species: 6
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 369 |
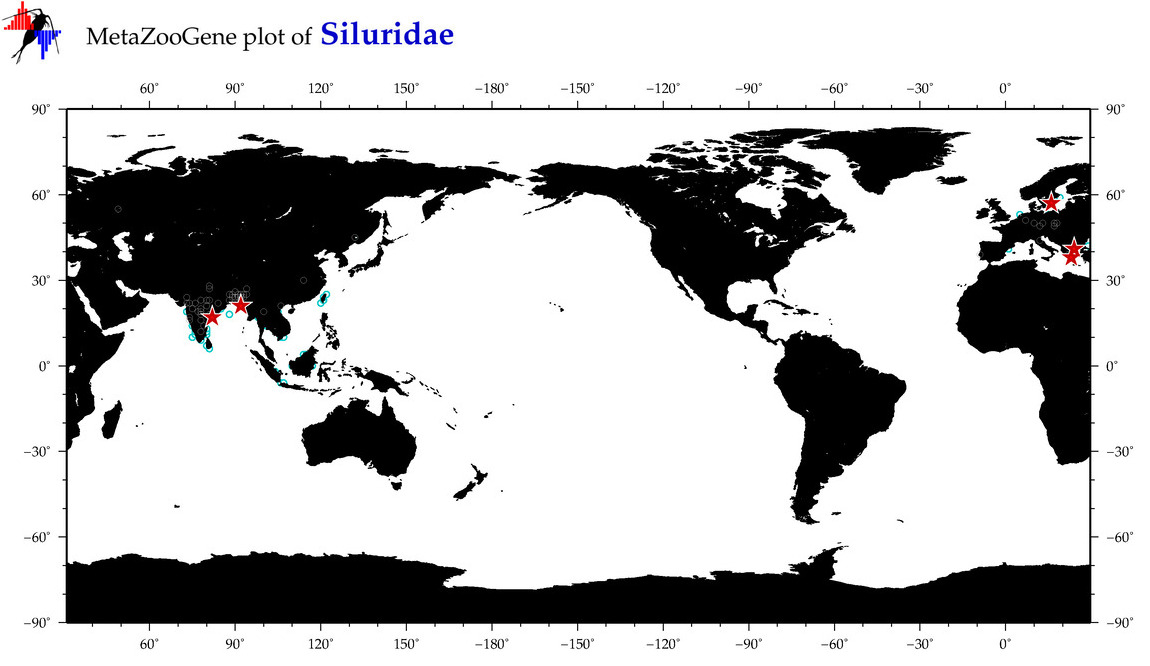 |
accepted
T5005956
R:1:1:1:1 |
| ⋄ ⋄ Kryptopterus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
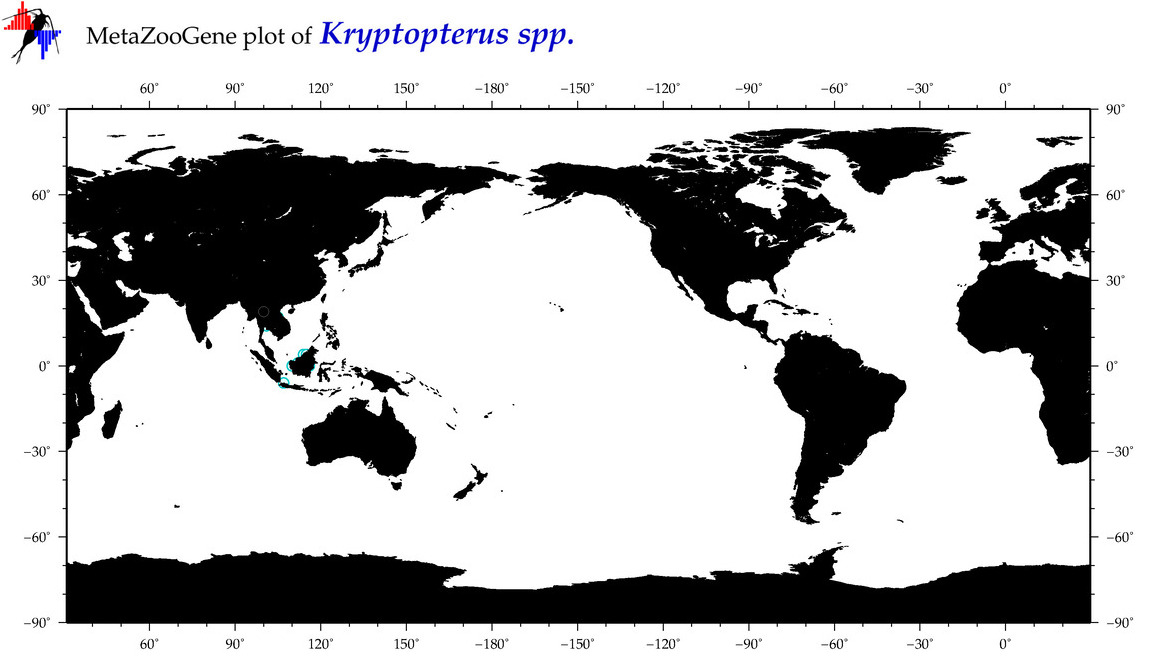 |
accepted
T5006867
R:0:1:1:0 |
| ⋄ ⋄ Ompok |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 176 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 176 |
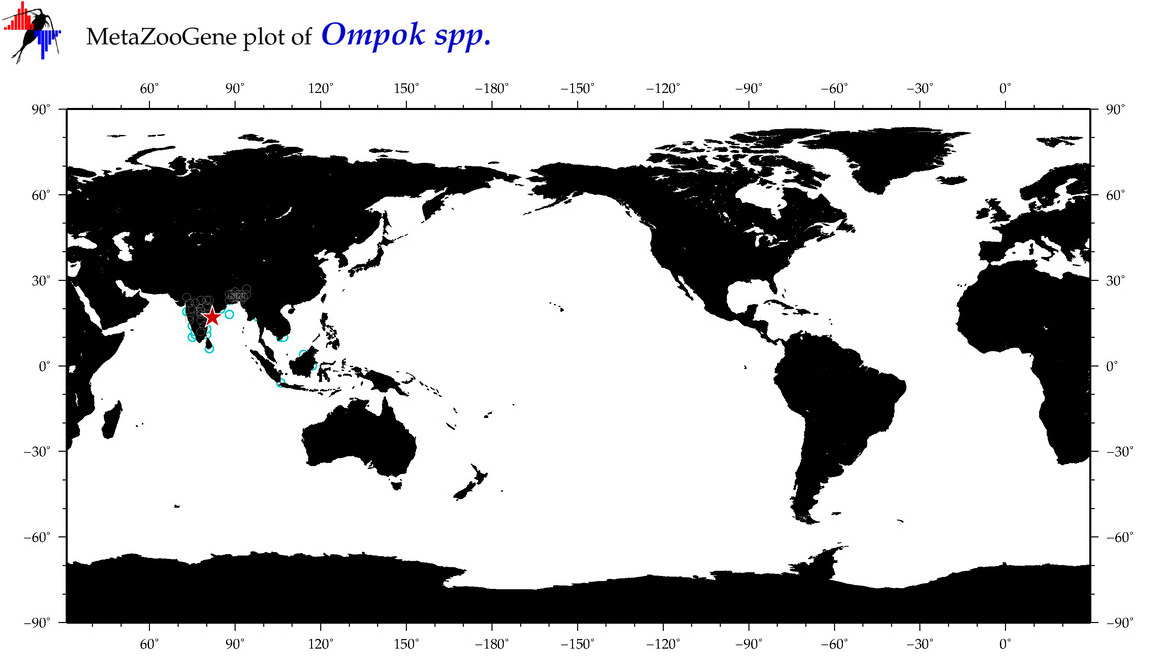 |
accepted
T5007028
R:0:1:1:1 |
| ⋄ ⋄ Silurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 79 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 79 |
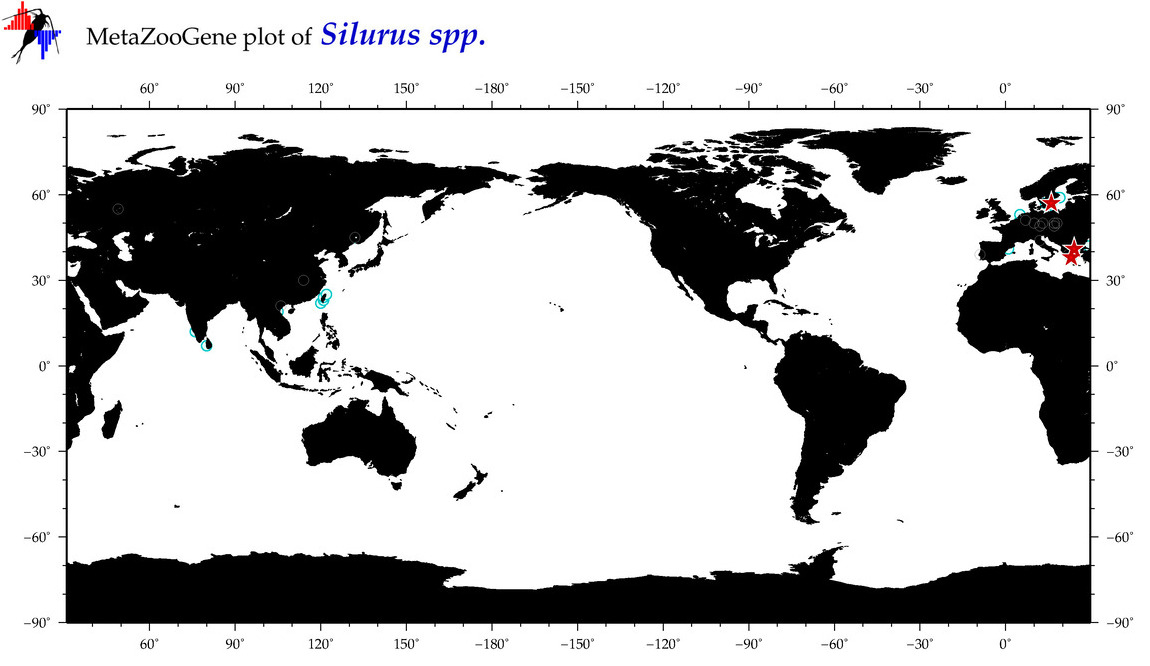 |
accepted
T5006260
R:0:1:1:0 |
| ⋄ ⋄ Wallago |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 111 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 111 |
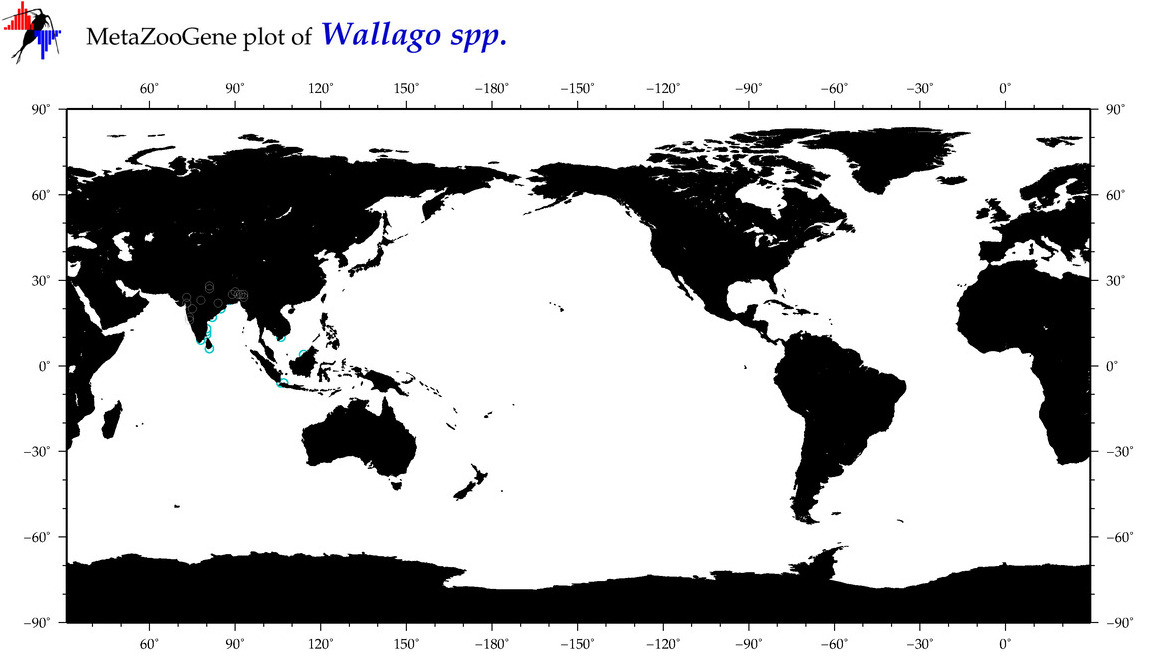 |
accepted
T5007409
R:0:1:1:0 |
| ⋄ Sisoridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 8
Sp. w/COI-Barcodes: 8
Total COI-Barcodes: 221 |
Total Species: 8
Sp. w/COI-Barcodes: 8
Total COI-Barcodes: 221 |
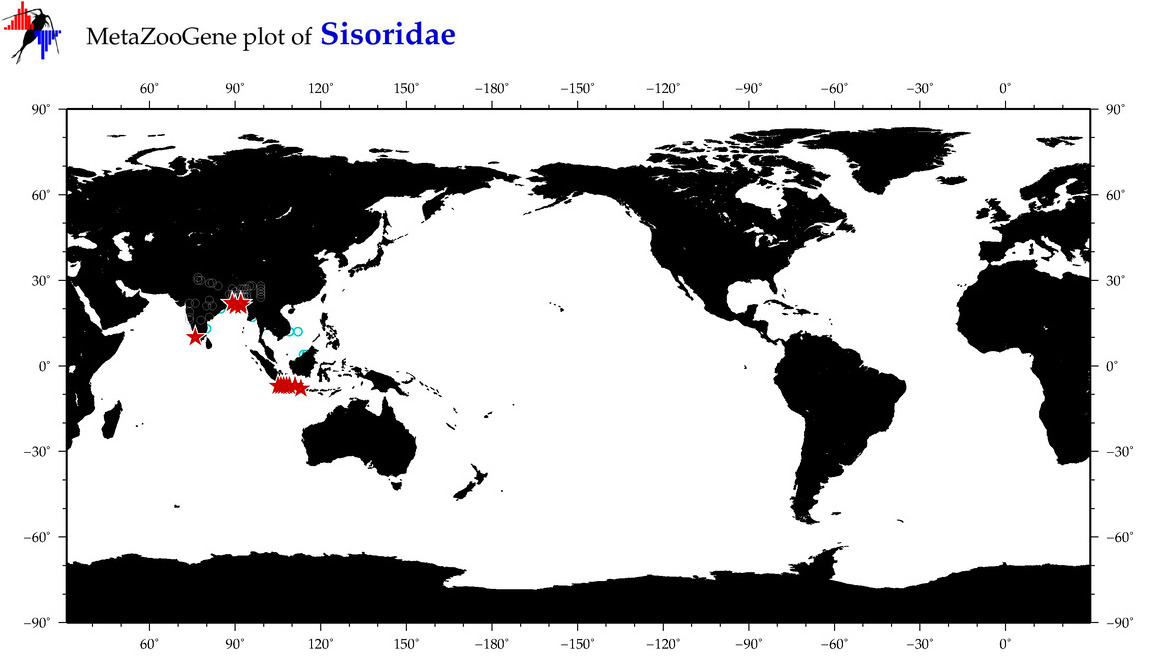 |
accepted
T5006023
R:0:1:1:0 |
| ⋄ ⋄ Glyptosterninae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 45 |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 45 |
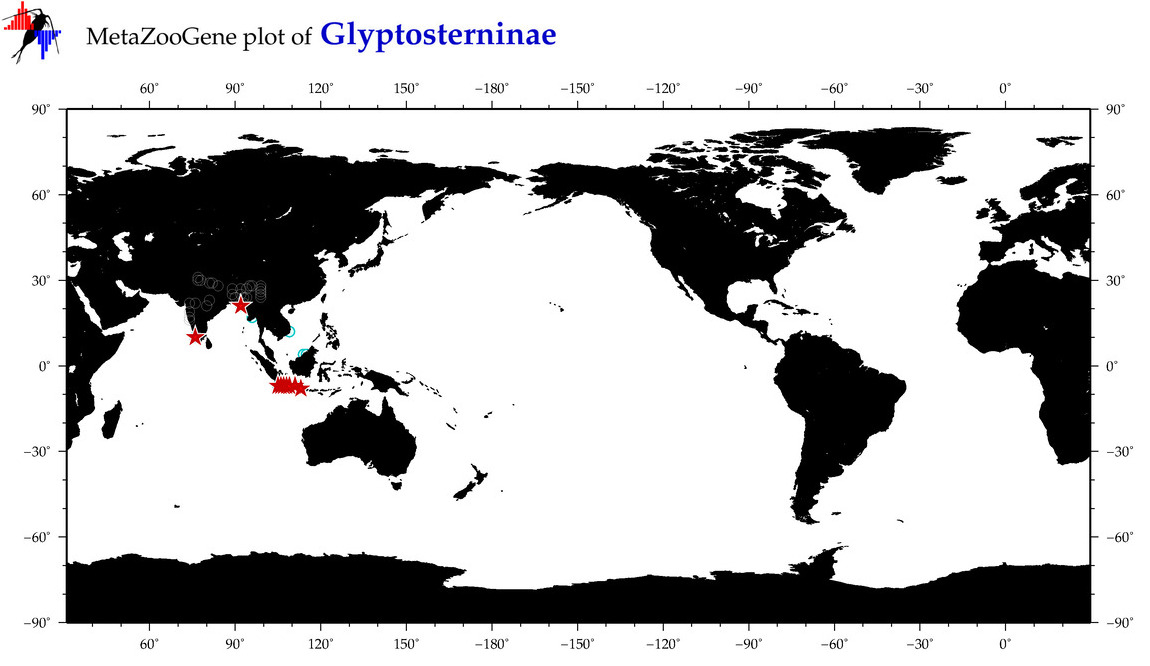 |
accepted
T5006145
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Glyptothorax |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 45 |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 45 |
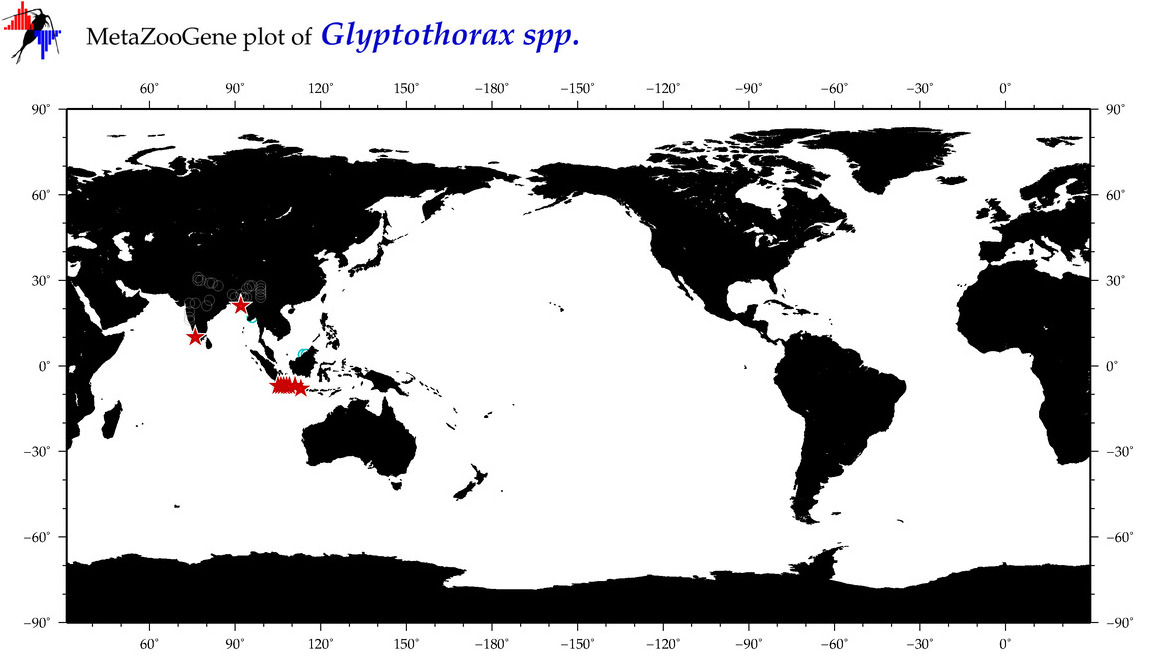 |
accepted
T5006749
R:0:1:1:0 |
| ⋄ ⋄ Sisorinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 176 |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 176 |
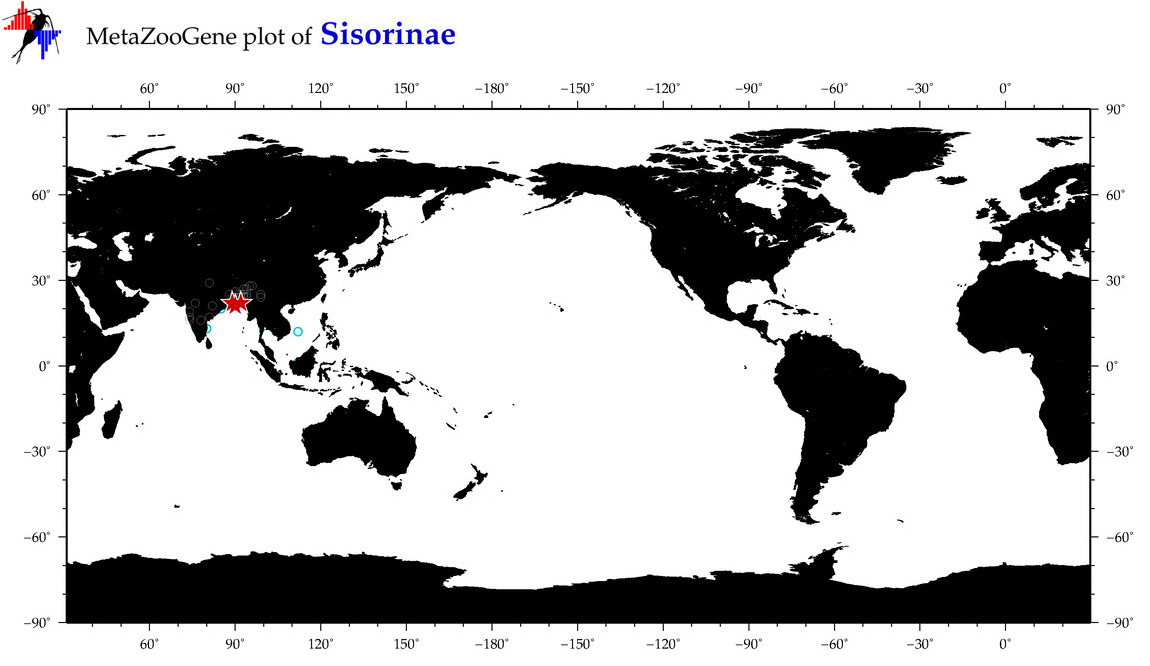 |
accepted
T5006146
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Bagarius |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 77 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 77 |
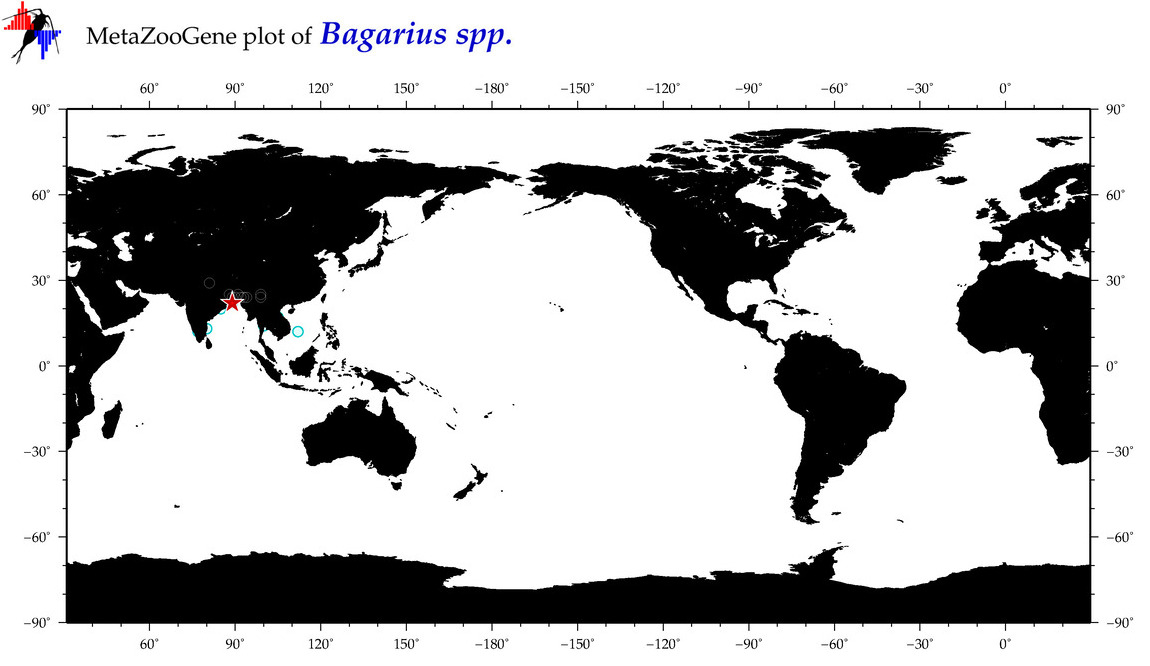 |
accepted
T5006507
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Gagata |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 99 |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 99 |
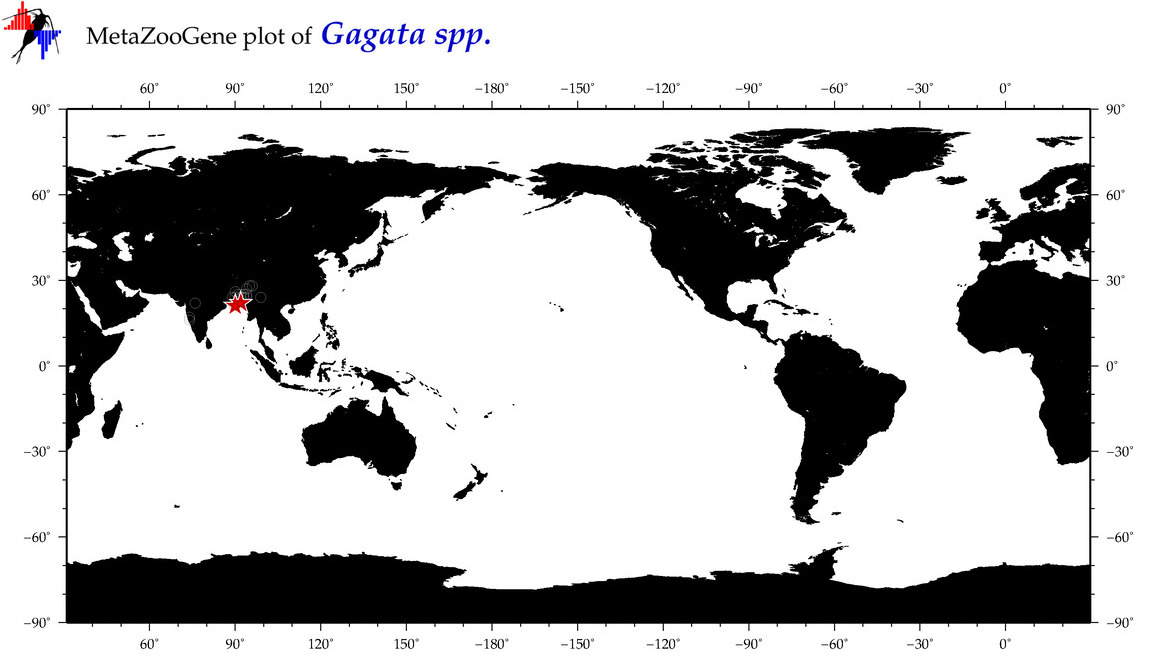 |
accepted
T5006730
R:0:1:1:0 |
| ⋄ Trichomycteridae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
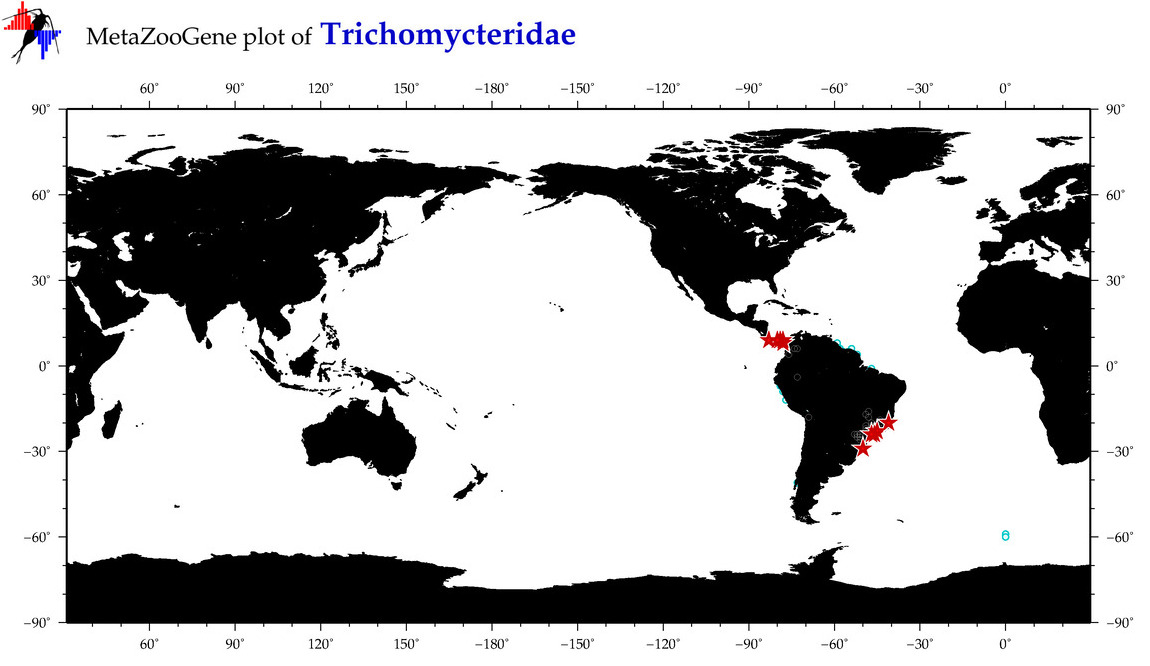 |
accepted
T5005997
R:0:1:1:0 |
| ⋄ ⋄ Tridentinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006193
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Potamoglanis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5030014
R:0:1:0:0 |