Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2023-m07-15
2023-Jul-20 |
| Allosmerus elongatus |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 4
16S = 4
18S = 0
28S = 0
|
COI = 15
12S = 4
16S = 4
18S = 0
28S = 0
|
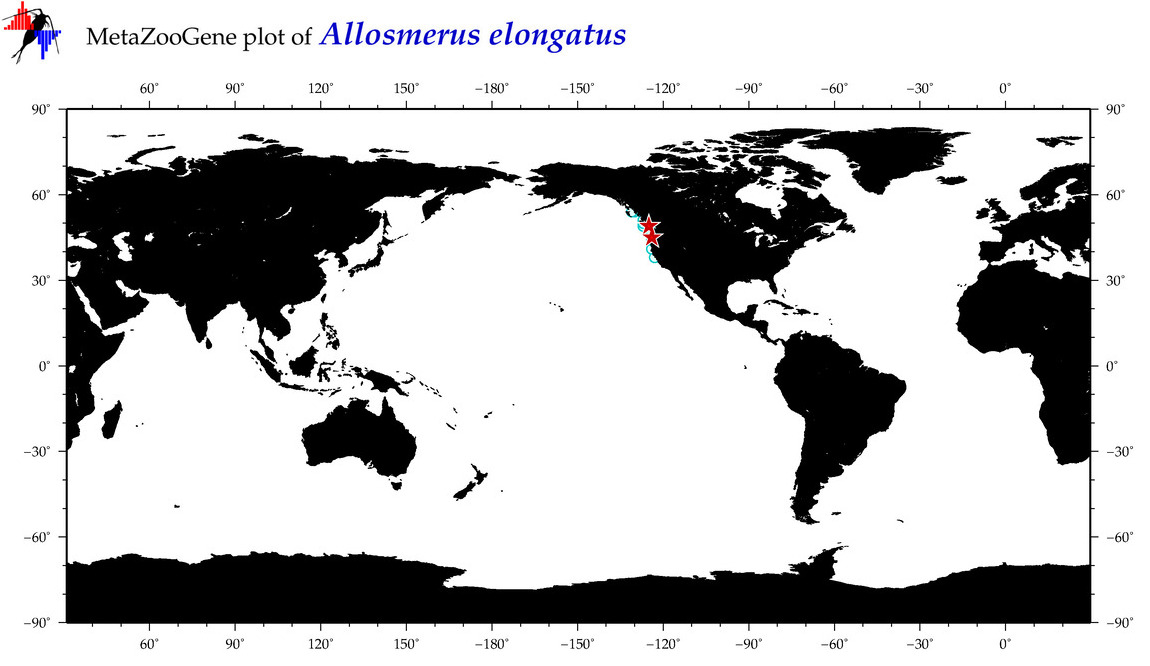 |
accepted
T5003067
R:1:0:0:0 |
| Hypomesus japonicus |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 6
16S = 8
18S = 0
28S = 0
|
COI = 15
12S = 6
16S = 8
18S = 0
28S = 0
|
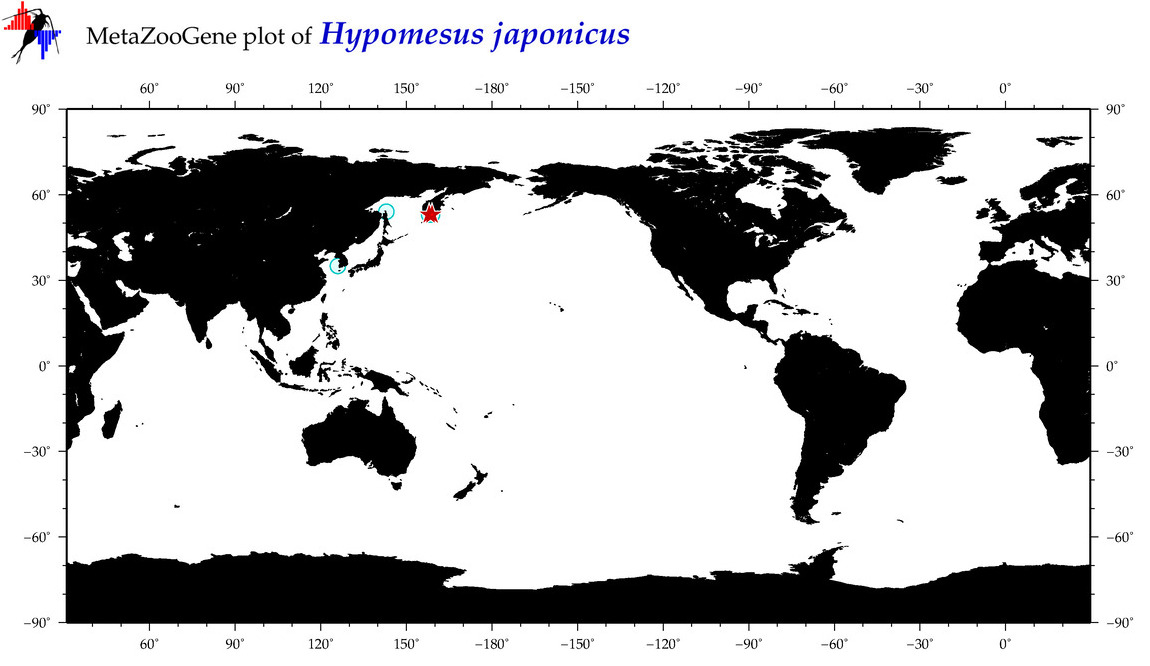 |
accepted
T5004299
R:1:1:1:0 |
| Hypomesus olidus |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 145
12S = 6
16S = 9
18S = 0
28S = 0
|
COI = 145
12S = 6
16S = 9
18S = 0
28S = 0
|
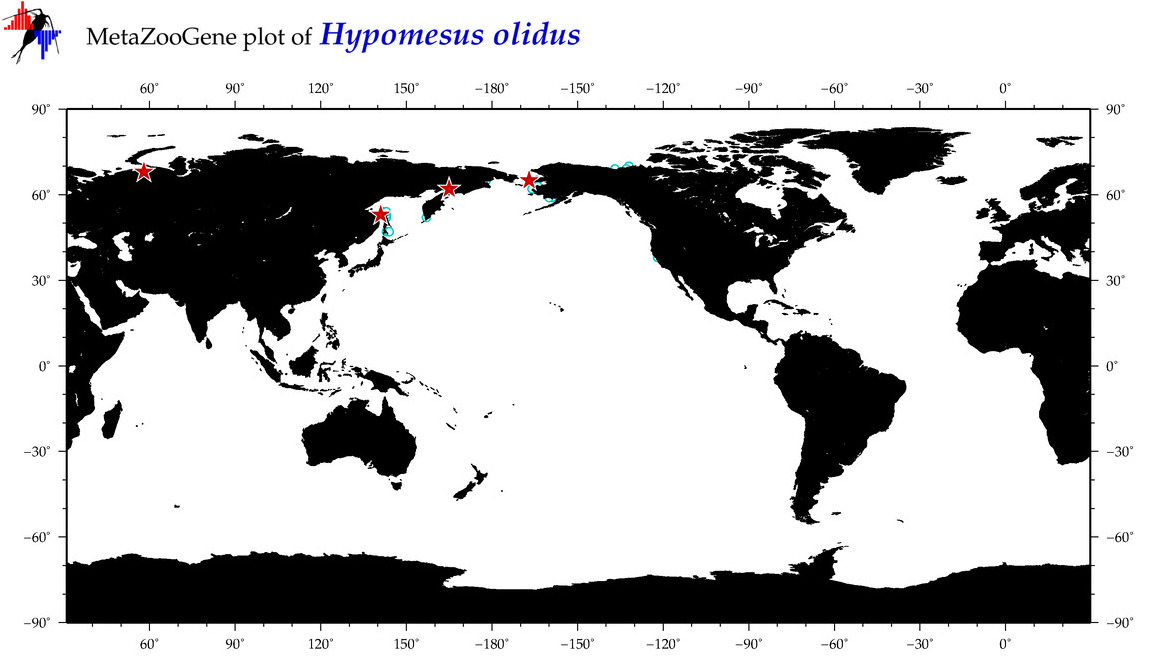 |
accepted
T5004301
R:1:1:1:0 |
| Hypomesus pretiosus |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 9
16S = 19
18S = 0
28S = 0
|
COI = 9
12S = 9
16S = 19
18S = 0
28S = 0
|
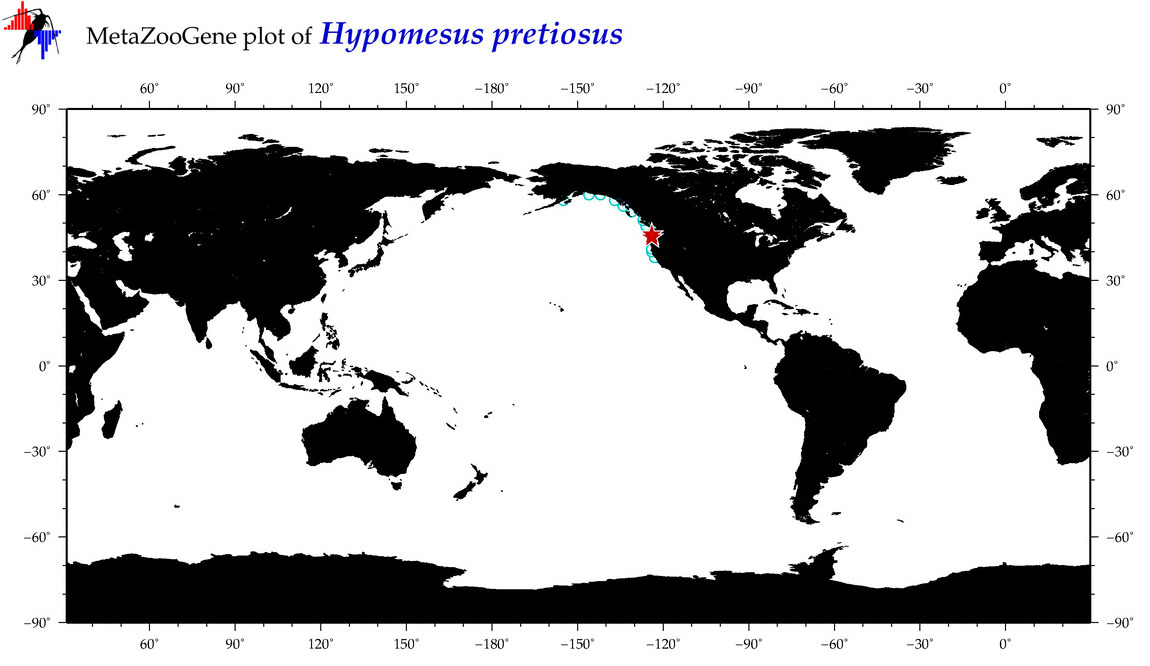 |
accepted
T5001822
R:1:1:0:0 |
| Leptochilichthys agassizii |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 2
12S = 2
16S = 1
18S = 0
28S = 0
|
no map |
accepted
T5021535
R:1:0:0:0 |
| Leptochilichthys microlepis |
Species
(6) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021536
R:1:0:0:0 |
| Leptochilichthys pinguis |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021537
R:1:0:0:0 |
| Mallotus villosus |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 59
12S = 8
16S = 11
18S = 0
28S = 0
|
COI = 59
12S = 8
16S = 11
18S = 0
28S = 0
|
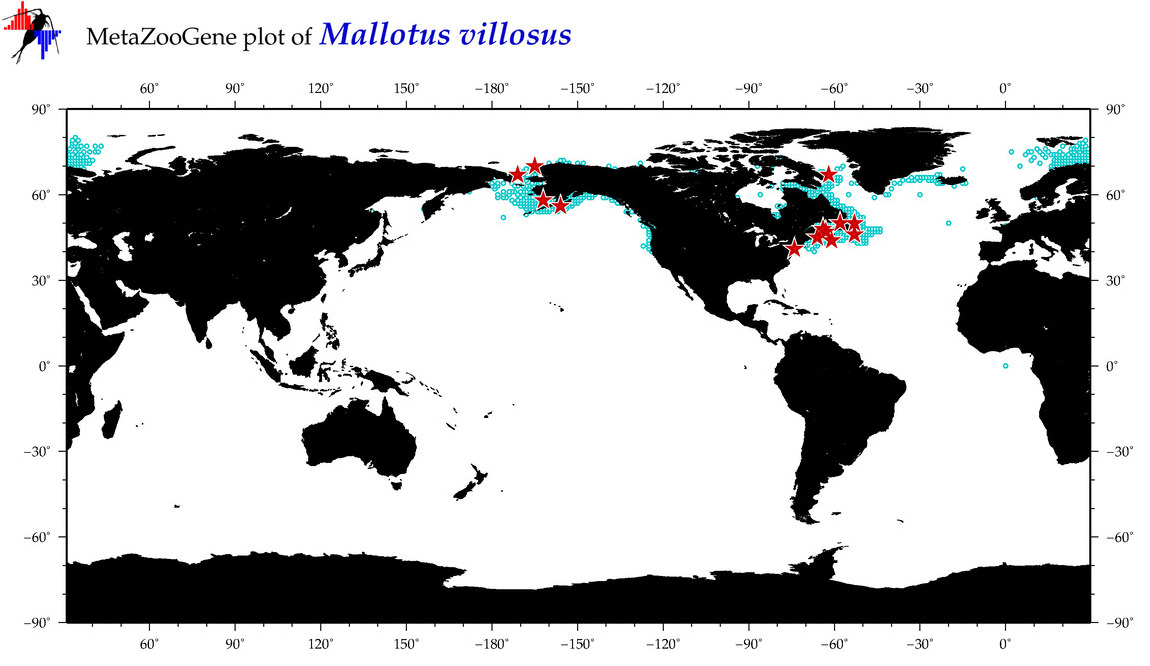 |
accepted
T5000868
R:1:1:1:0 |
| Neosalangichthys ishikawae |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 6
16S = 5
18S = 0
28S = 3
|
COI = 0
12S = 6
16S = 5
18S = 0
28S = 3
|
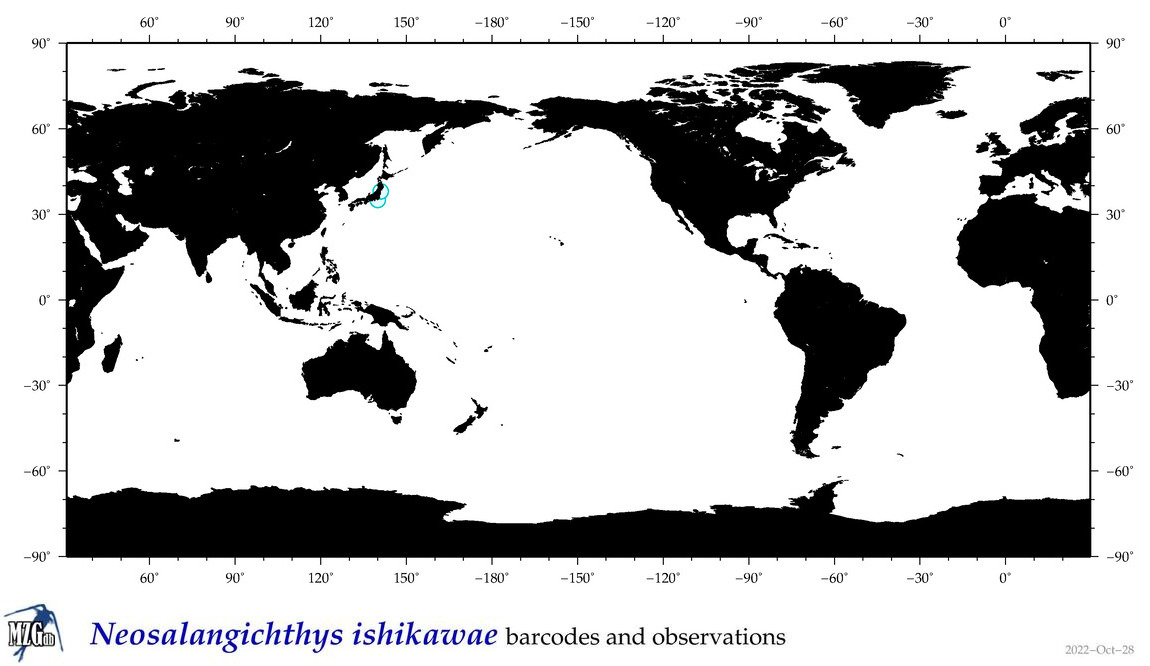 |
accepted
T5028349
R:1:1:1:0 |
| Neosalanx anderssoni |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 7
16S = 8
18S = 0
28S = 5
|
COI = 10
12S = 7
16S = 8
18S = 0
28S = 5
|
 |
accepted
T5010658
R:1:1:1:0 |
| Neosalanx brevirostris |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 11
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 11
|
 |
accepted
T5010660
R:1:1:1:0 |
| Neosalanx jordani |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 4
16S = 5
18S = 0
28S = 5
|
COI = 3
12S = 4
16S = 5
18S = 0
28S = 5
|
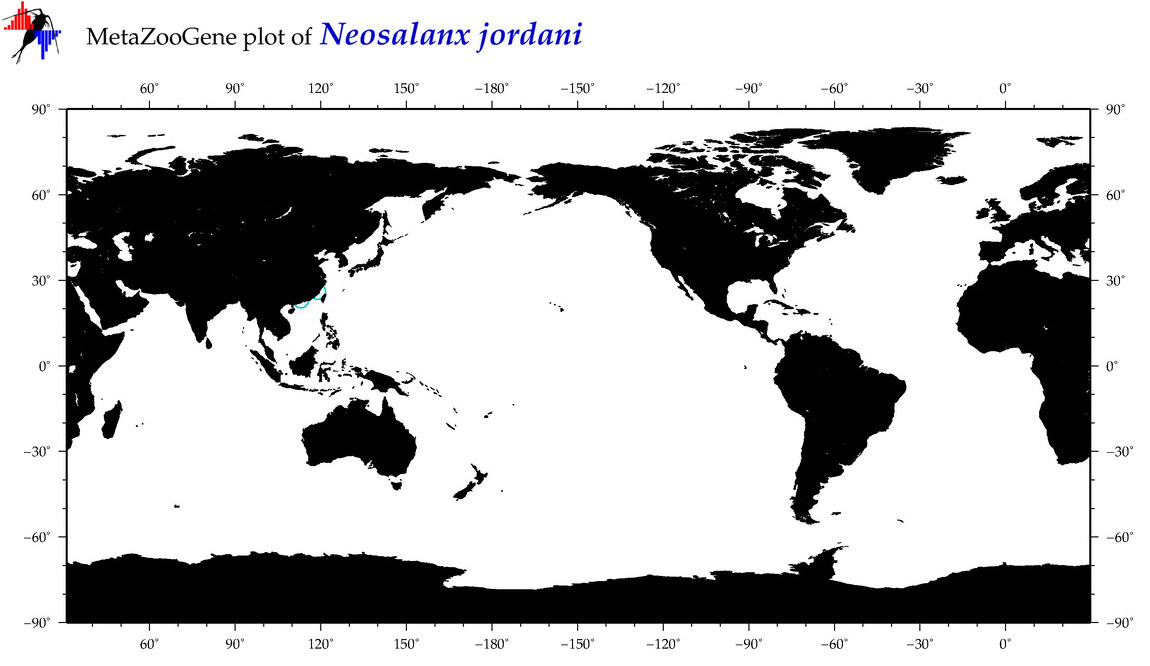 |
accepted
T5010661
R:1:1:1:0 |
| Neosalanx reganius |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
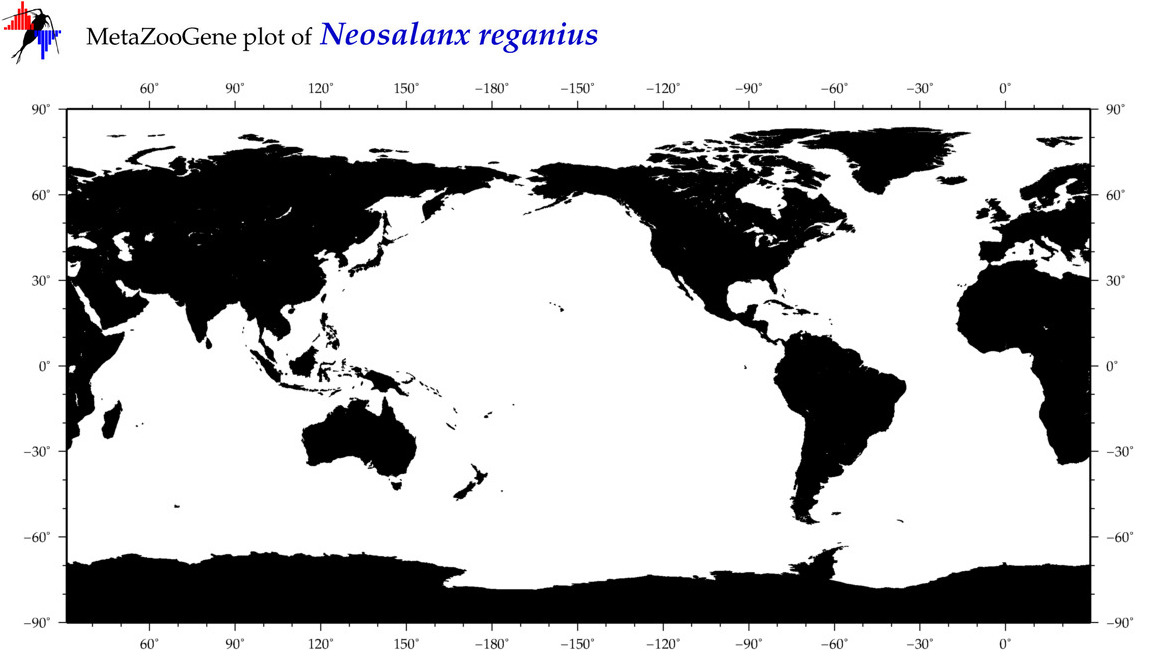 |
accepted
T5010664
R:1:1:1:0 |
| Osmerus dentex |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5028399
R:1:1:1:0 |
| Osmerus eperlanus |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 35
12S = 7
16S = 10
18S = 0
28S = 1
|
COI = 35
12S = 7
16S = 10
18S = 0
28S = 1
|
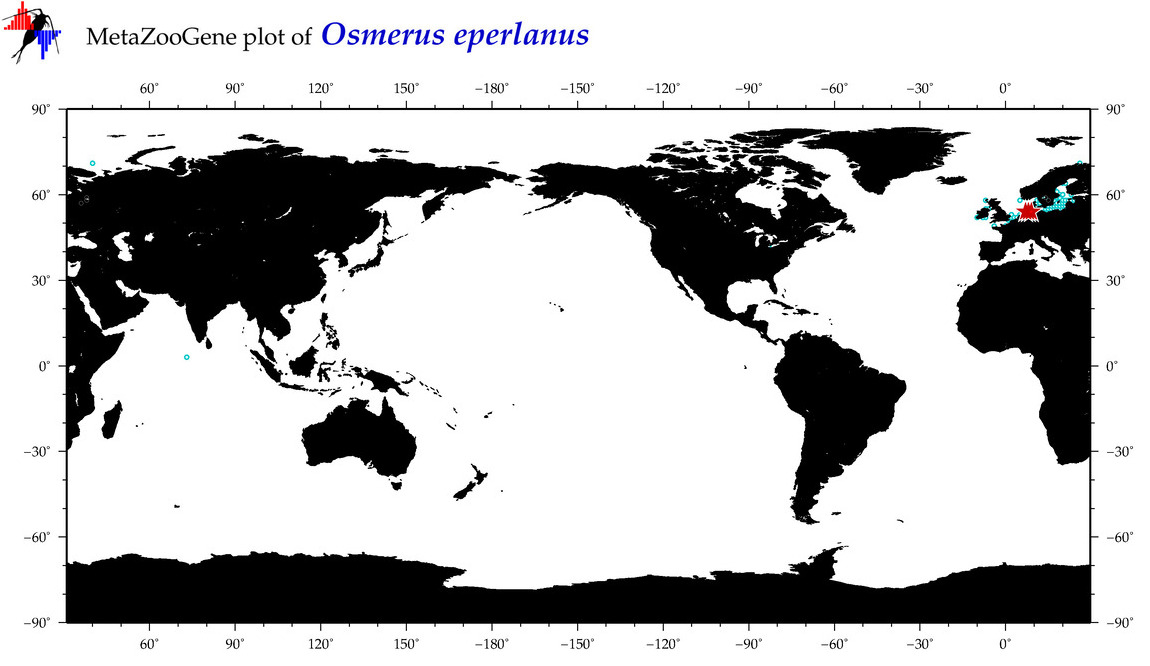 |
accepted
T5010980
R:1:1:1:0 |
| Osmerus mordax |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 129
12S = 15
16S = 18
18S = 1
28S = 1
|
COI = 129
12S = 15
16S = 18
18S = 1
28S = 1
|
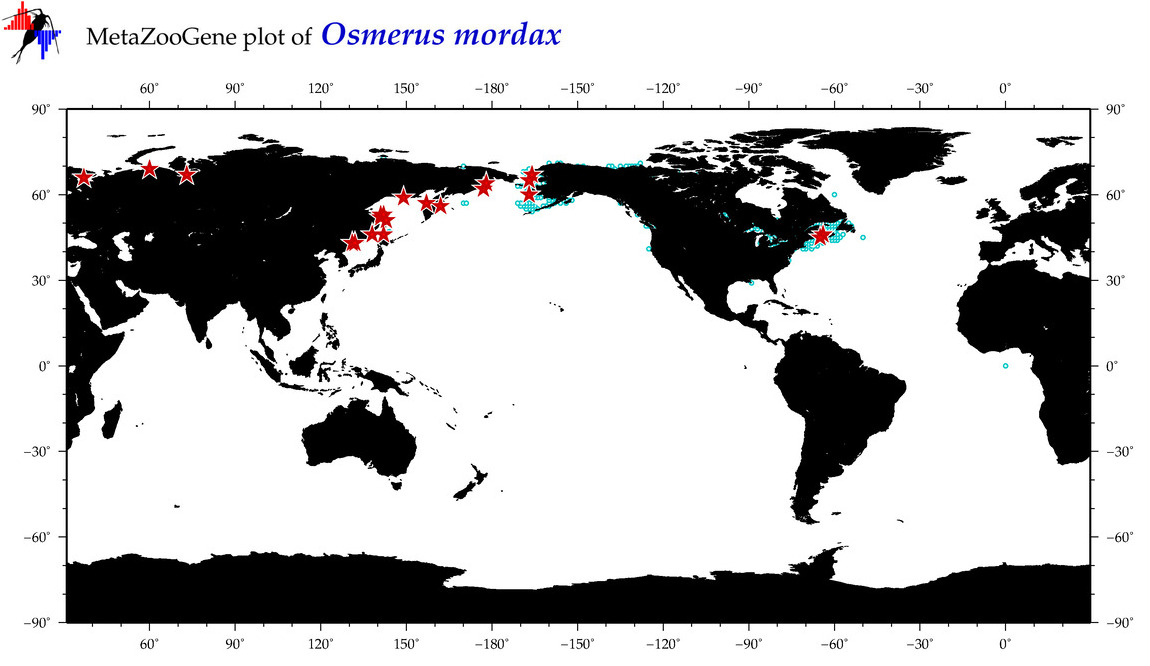 |
accepted
T5010981
R:1:1:1:0 |
| Plecoglossus altivelis |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 19
16S = 11
18S = 1
28S = 1
|
COI = 15
12S = 19
16S = 11
18S = 1
28S = 1
|
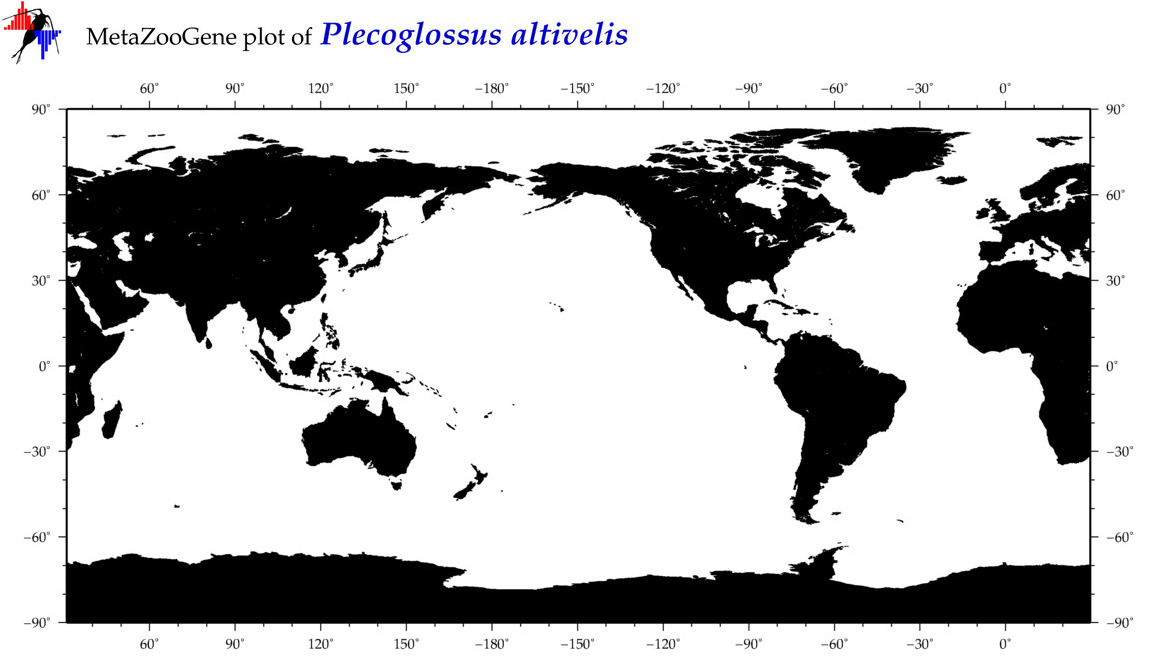 |
accepted
T5005060
R:1:1:1:0 |
| Protosalanx hyalocranius |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 17
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5011605
R:1:1:1:0 |
| Prototroctes maraena |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 4
16S = 7
18S = 1
28S = 0
|
COI = 4
12S = 4
16S = 7
18S = 1
28S = 0
|
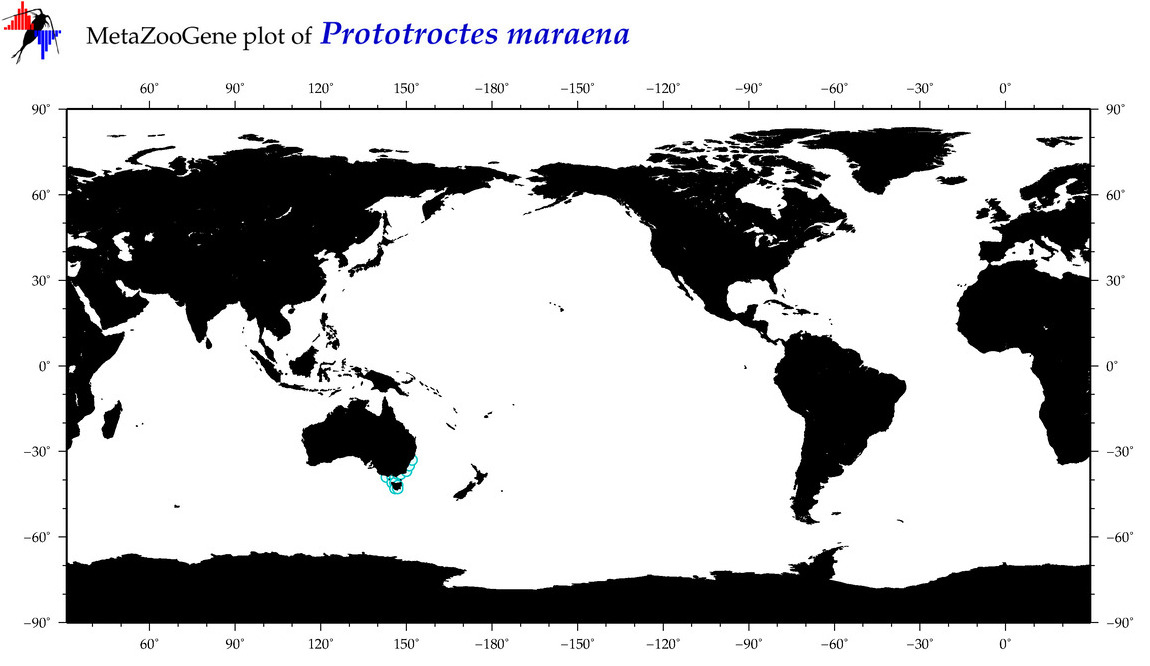 |
accepted
T5011606
R:1:1:1:0 |
| Retropinna retropinna |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 8
18S = 0
28S = 0
|
COI = 2
12S = 1
16S = 8
18S = 0
28S = 0
|
no map |
accepted
T5024896
R:1:1:1:0 |
| Retropinna tasmanica |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 2
16S = 1
18S = 2
28S = 0
|
COI = 1
12S = 2
16S = 1
18S = 2
28S = 0
|
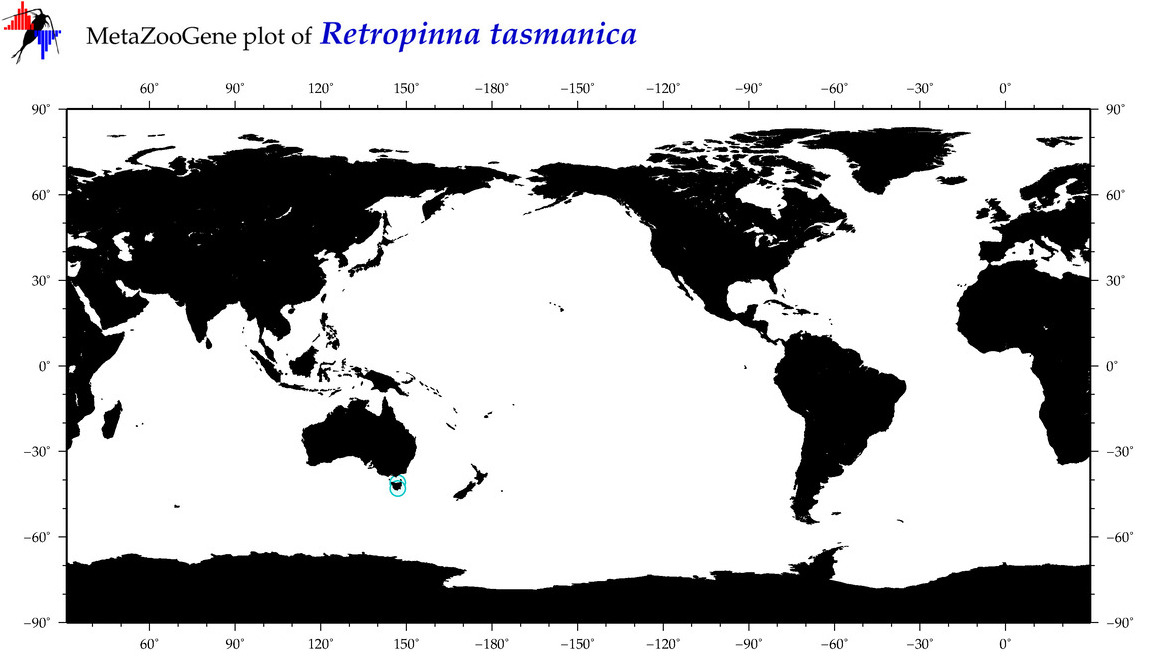 |
accepted
T5011873
R:1:1:1:0 |
| Salangichthys ishikawae |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
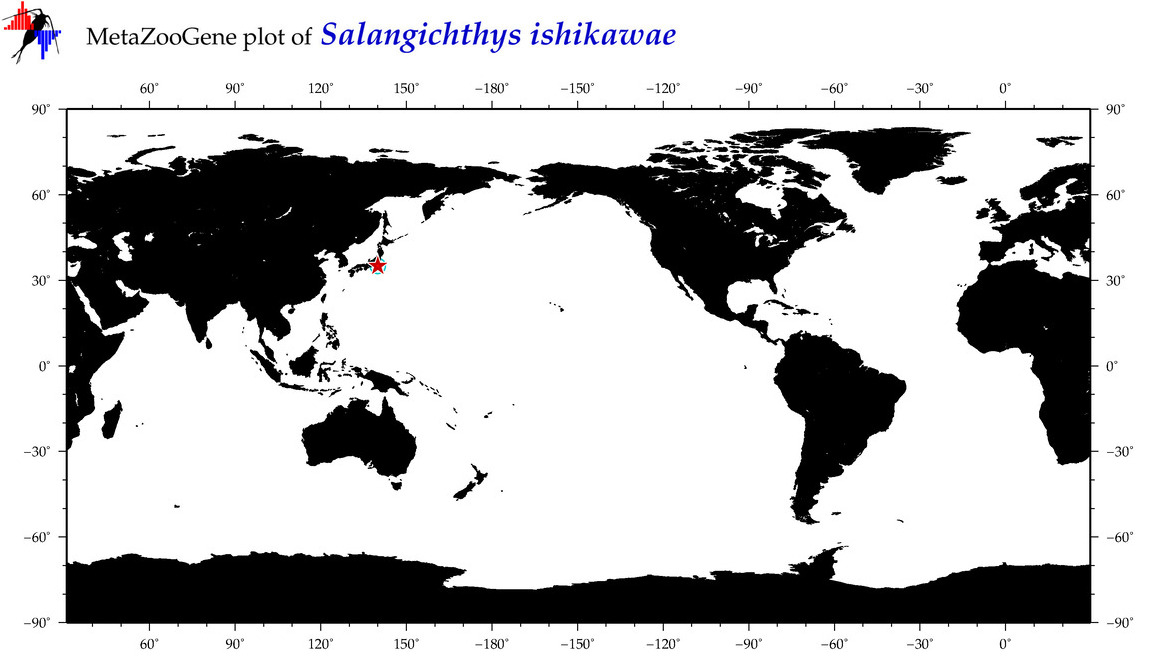 |
accepted
T5011993
R:1:1:1:0 |
| Salangichthys microdon |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 8
16S = 8
18S = 0
28S = 3
|
COI = 9
12S = 8
16S = 8
18S = 0
28S = 3
|
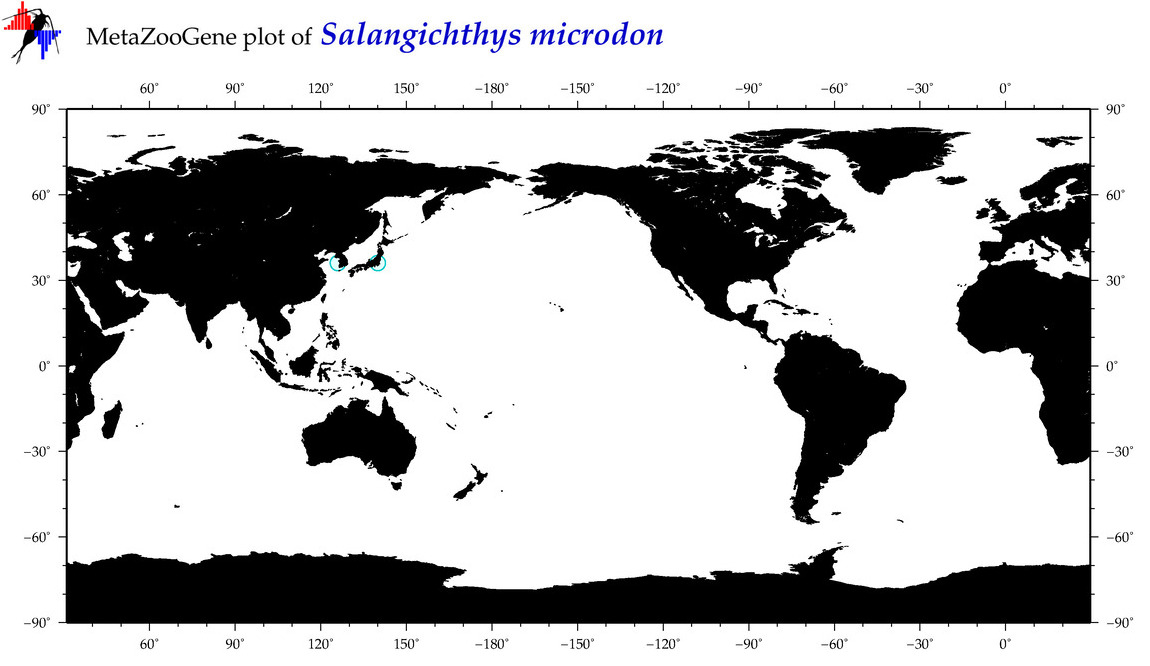 |
accepted
T5011994
R:1:1:1:0 |
| Salanx ariakensis |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 6
16S = 7
18S = 0
28S = 3
|
COI = 14
12S = 6
16S = 7
18S = 0
28S = 3
|
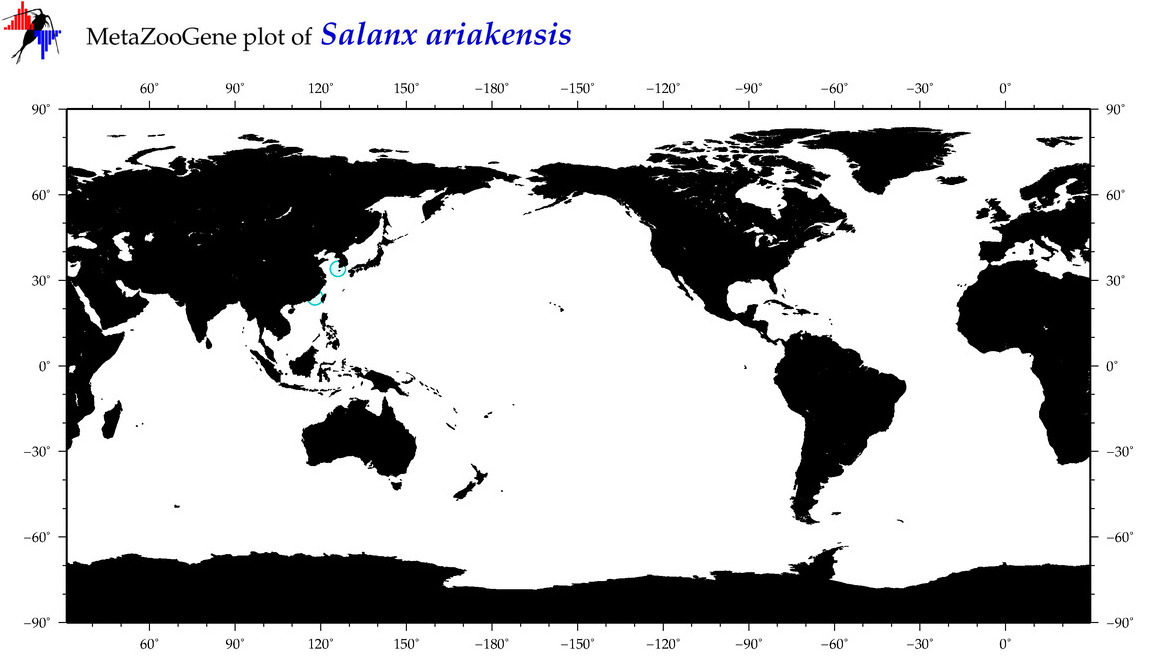 |
accepted
T5011995
R:1:1:1:0 |
| Salanx chinensis |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 5
16S = 4
18S = 0
28S = 0
|
COI = 11
12S = 5
16S = 4
18S = 0
28S = 0
|
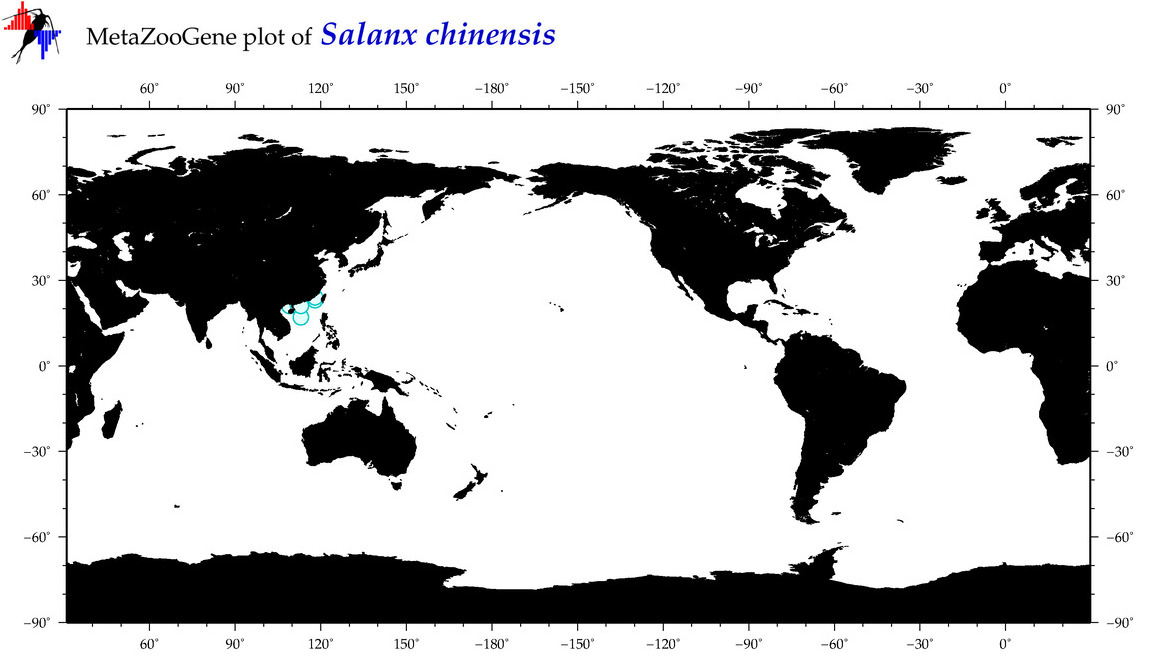 |
accepted
T5011996
R:1:1:1:0 |
| Salanx cuvieri |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 5
16S = 5
18S = 0
28S = 3
|
COI = 12
12S = 5
16S = 5
18S = 0
28S = 3
|
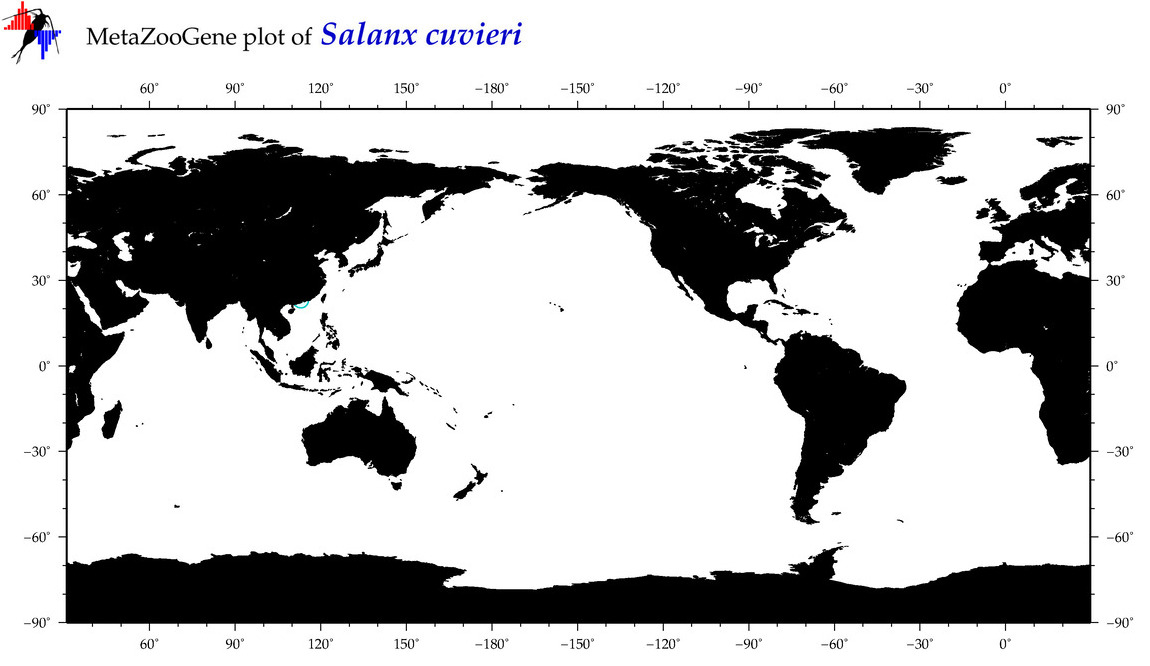 |
accepted
T5011997
R:1:1:1:0 |
| Salanx prognathus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 3
|
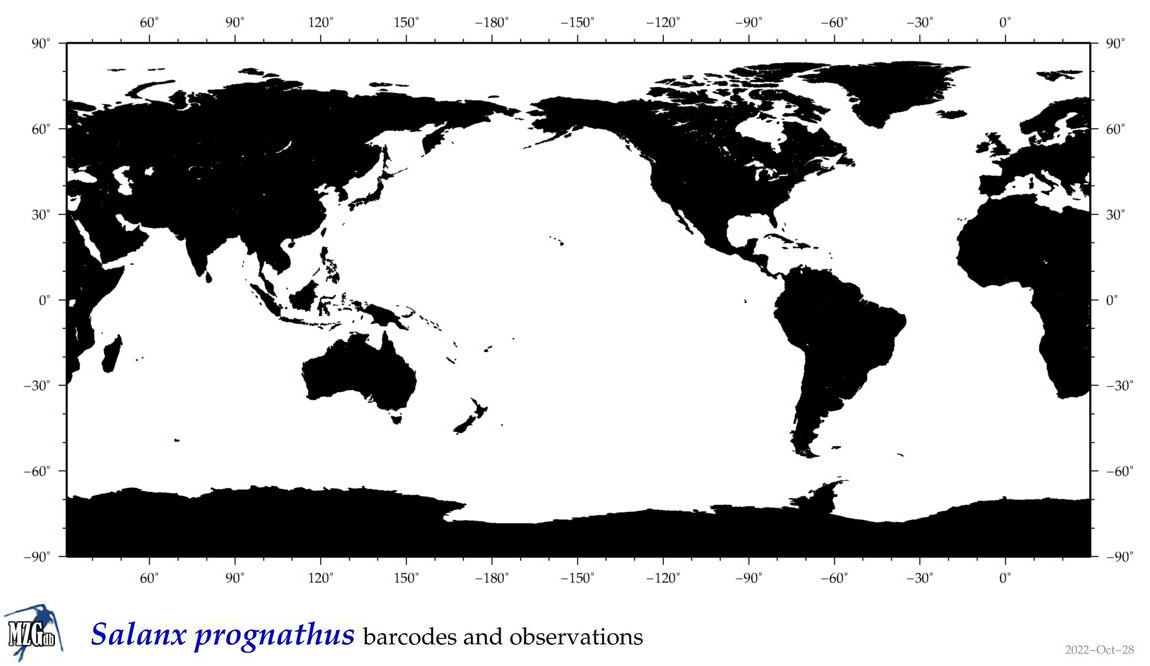 |
accepted
T5028615
R:1:1:1:0 |
| Spirinchus lanceolatus |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 4
16S = 7
18S = 2
28S = 0
|
COI = 0
12S = 4
16S = 7
18S = 2
28S = 0
|
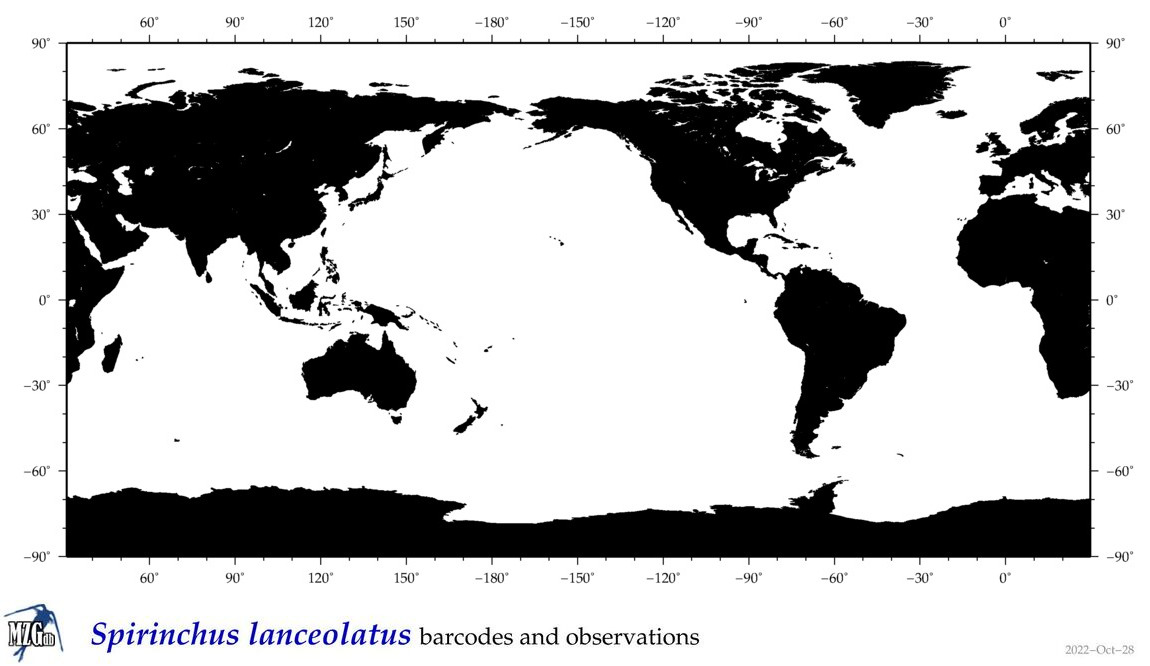 |
accepted
T5028682
R:1:1:1:0 |
| Spirinchus starksi |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 4
16S = 6
18S = 0
28S = 0
|
COI = 8
12S = 4
16S = 6
18S = 0
28S = 0
|
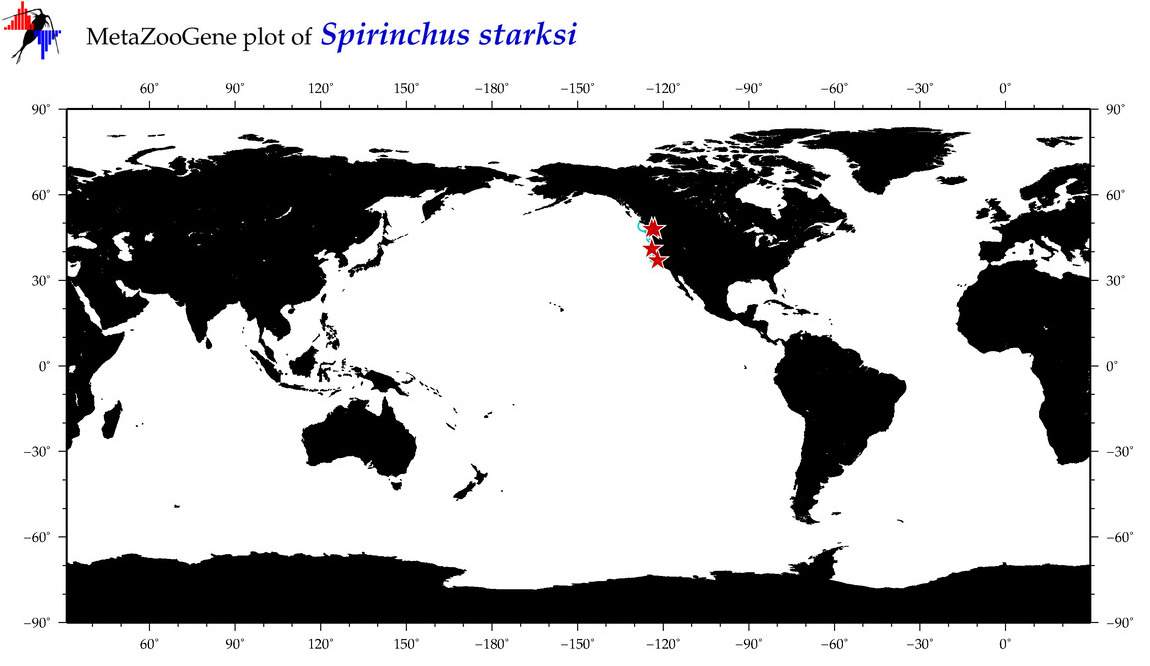 |
accepted
T5005597
R:1:0:0:0 |
| Spirinchus thaleichthys |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 9
16S = 7
18S = 0
28S = 0
|
COI = 8
12S = 9
16S = 7
18S = 0
28S = 0
|
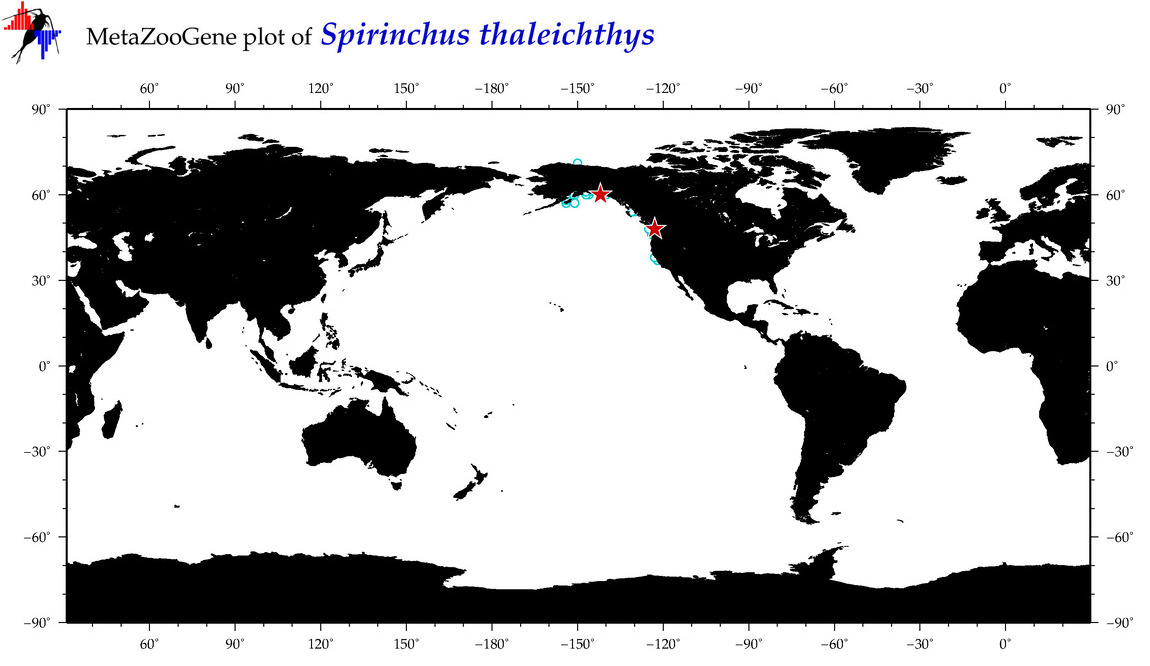 |
accepted
T5005598
R:1:1:1:0 |
| Stokellia anisodon |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 2
16S = 7
18S = 0
28S = 0
|
COI = 1
12S = 2
16S = 7
18S = 0
28S = 0
|
 |
accepted
T5012381
R:1:1:1:0 |
| Thaleichthys pacificus |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 37
12S = 7
16S = 16
18S = 0
28S = 0
|
COI = 37
12S = 7
16S = 16
18S = 0
28S = 0
|
 |
accepted
T5000004
R:1:1:1:0 |